Abstract
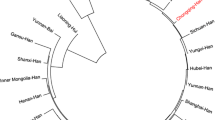
The genetic polymorphism of 17 autosomal short tandem repeat (STR) loci included in the PowerPlex® 18D System was evaluated from 638 unrelated healthy Han individuals of Meizhou in Guangdong Province, Southern China. The values of combined power of discrimination (CPD) and the combined probability of exclusion (CPE) were observed as 0.999999999999999 and 0.999999933. Penta E showed the highest values of polymorphism information content (0.9073), expected heterozygosity (0.9147), and observed heterozygosity (0.9373), whereas TPOX showed the lowest (0.5373, 0.6035, and 0.6082). Furthermore, both of the PCA plot and neighbor-joining phylogenetic tree showed that the Meizhou Hakka population has a relatively close genetic relationship with the Guangdong Han population. The results showed that most of these 17 autosomal STR loci were highly informative and can be effective for forensic individual identification and paternity testing in Meizhou Hakka population.
Similar content being viewed by others
References
Wu S (1995) The origin and development theory of Hakka in Southern Song Dynasty. Fudan university journal of social sciences 5:108–113
Zhang T (1997) Hakka formed in the Southern Song Dynasty. Academic Journal of Zhongzhou 2:119–124
Zhu J (2014) National treasure. People’s Art Publishing House, Beijing
Tan Y (2008) Hakka illustrated record. Lingnan Art Publishing House, Guangdong
Weispfenning R et al (2011) Doing more with less: implementing direct amplification with the PowerPlex® 18D system. Forensic Science International Genetics Supplement 3(1):e409–e410
Oostdik, et al., Developmental validation of the PowerPlex (R) 18D System, a rapid STR;multiplex for analysis of reference samples. Forensic Science International Genetics, 2013. 7(1): p. 129–135
TerebaA, Tools for analysis of population statistics; Promaga Corporation[J]. 1999
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Naoko T, Masatoshi N, Koichiro T (2010) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Molecular Biology & Evolution 27(4):747–752
Hammer Ø, Dat H, Pd R Past: paleontological statistics software package for education and data analysis. Palaeontol Electron
Du W et al (2018) Genetic polymorphisms of 32 Y-STR loci in Meizhou Hakka population. Int J Legal Med 2–3:1–2
Acknowledgments
The authors are very grateful to all sample donors for their contributions to this work and all those who helped with sample collection.
Funding
This study was funded by the National Natural Science Foundation of China (81971798) and the Natural Science Foundation of Guangdong Province (2019A15150).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Informed consents were obtained before sample collection.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Han, X., Shen, A., Yao, T. et al. Genetic diversity of 17 autosomal STR loci in Meizhou Hakka population. Int J Legal Med 135, 443–444 (2021). https://doi.org/10.1007/s00414-020-02253-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-020-02253-9