Abstract
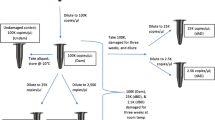
Human remains can be severely affected by the environment, and the DNA may be damaged, degraded, and/or inhibited. In this study, a DNA sample (at 1 ng DNA target input in triplicate) was spiked with five concentrations of five inhibitors (humic acid, melanin, hematin, collagen, and calcium) and sequenced with both the HID-Ion AmpliSeq™ Library Kit and ID panel on the Ion PGM™ System and the ForenSeq DNA Signature Prep Kit on the MiSeq FGx™. The objective of this study was to compare the baseline tolerance of the two sequencing chemistries and platforms to common inhibitors encountered in human remains recovered from missing person cases. The two chemistries generally were comparable but not always susceptible to the same inhibitors or at the same capacity. The HID-Ion AmpliSeq™ Library Kit and ID panel and the ForenSeq DNA Signature Prep Kit both were susceptible to humic acid, melanin, and collagen; however, the ForenSeq kit showed greater inhibition to melanin and collagen than the AmpliSeq™ kit. In contrast, the ForenSeq kit was resistant to the effects of hematin and calcium, whereas the AmpliSeq™ kit was highly inhibited by hematin. Short tandem repeats (STRs) and single nucleotide polymorphisms (SNPs) showed the same trend among inhibitors when using the ForenSeq kit. Generally, locus read depth, heterozygote allele balance, and the numbers of alleles typed were inversely correlated with increasing inhibitor concentration. The larger STR loci were affected more so by the presence of inhibitors compared to smaller STR amplicons and SNP loci. Additionally, it does not appear that sequence noise is affected by the inhibitors. The noise percentage, however, does increase as the inhibitor concentration increases, due to the decrease in locus read depth and not likely because of chemistry effects.










Similar content being viewed by others
References
Ritter N (2007) Missing persons and unidentified remains: the nation’s silent mass disaster. NIJ Journal 256:2–7
Kovras I, Robins S (2016) Death as the border: managing missing migrants and unidentified bodies at the EU’s Mediterranean frontier. Polit Geogr 55:40–49
Dhanardhono T, Wulandari N, Bhima SKL, Ahmad HJ, Widodo PT (2013) DNA profiling of disaster victim identification in Trenggalek shipwreck case. For Sci Int Genet Supplement Series 4:e5–e6
Barbisin M, Fang R, O’Shea CE, Calandro LM, Furtado MR, Shewale JG (2009) Developmental validation of the Quantifiler® duo DNA quantification kit for simultaneous quantification of total human and human male DNA and detection of PCR inhibitors in biological samples. J Forensic Sci 54:305–319
Mulero JJ, Chang CW, Lagace RE, Wang DY, Bas JL, McMahon TP, Hennessy LK (2008) Development and validation of the AmpFℓSTR® MiniFilerTM PCR amplification kit: a miniSTR multiplex for the analysis of degraded and/or PCR inhibited DNA. J Forensic Sci 53:838–852
Thompson RE, Duncan G, McCord BR (2014) An investigation of PCR inhibition using Plexor®-based quantitative PCR and short tandem repeat amplification. J Forensic Sci 59:1517–1529
Opel KL, Chung D, McCord BR (2010) A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci 55:25–33
Collins PJ, Hennessy LK, Leibelt CS, Roby RK, Reeder DJ, Foxall PA (2004) Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: the AmpFlSTR Identifiler PCR amplification kit. J Forensic Sci 49:1265–1277
Krenke BE, Tereba A, Anderson SJ, Buel E, Culhane S, Finis CJ, Tomsey CS, Zachetti JM, Masibay A, Rabbach DR, Amiott EA, Sprecher CJ (2002) Validation of a 16-locus fluorescent multiplex system. J Forensic Sci 47:773–785
Martín P, de Simón LF, Luque G, Farfán MJ, Alonso A (2014) Improving DNA data exchange: validation studies on a single 6 dye STR kit with 24 loci. For Sci Int Genet 13C:68–78
Oostdik K, Lenz K, Nye J, Schelling K, Yet D, Bruski S, Strong J, Buchanan C, Sutton J, Linner J, Frazier N, Young H, Matthies L, Sage A, Hahn J, Wells R, Williams N, Price M, Koehler J, Staples M, Swango KL, Hill C, Oyerly K, Duke W, Katzilierakis L, Ensenberger MG, Bourdeau JM, Sprecher CJ, Krenke B, Storts DR (2014) Developmental validation of the PowerPlex((R)) fusion system for analysis of casework and reference samples: a 24-locus multiplex for new database standards. For Sci Int Genet 12:69–76
Lazaruk K, Walsh PS, Oaks F, Gilbert D, Rosenblum BB, Menchen S, Scheibler D, Wenz HM, Holt C, Wallin J (1998) Genotyping of forensic short tandem repeat (STR) systems based on sizing precision in a capillary electrophoresis instrument. Electrophoresis 19:86–93
Butler JM, Buel E, Crivellente F, McCord BR (2004) Forensic DNA typing by capillary electrophoresis using the ABI Prism 310 and 3100 genetic analyzers for STR analysis. Electrophoresis 25:1397–1412
Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N (2006) A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27:1713–1724
Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SL, Karoma NJ, Kungulilo S, Kim JJ, Lu RB, Odunsi A, Okonofua F, Parnas J, Schulz LO, Zhukova OV, Kidd JR (2006) Developing a SNP panel for forensic identification of individuals. For Sci Int Genet 164:20–32
Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, Shigeta R, Silva G, Patel PI, Belmont JW, Seldin MF (2005) Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet 118:382–392
Kayser M, Schneider PM (2009) DNA-based prediction of human externally visible characteristics in forensics: motivations, scientific challenges, and ethical considerations. For Sci Int Genet 3:154–161
Spichenok O, Budimlija ZM, Mitchell AA, Jenny A, Kovacevic L, Marjanovic D, Caragine T, Prinz M, Wurmbach E (2011) Prediction of eye and skin color in diverse populations using seven SNPs. For Sci Int Genet 5:472–478
Walsh S, Liu F, Wollstein A, Kovatsi L, Ralf A, Kosiniak-Kamysz A, Branicki W, Kayser M (2013) The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. For Sci Int Genet 7:98–115
Chakraborty R, Stivers DN, Su B, Zhong Y, Budowle B (1999) The utility of short tandem repeat loci beyond human identification: implications for development of new DNA typing systems. Electrophoresis 20:1682–1696
Zeng X, King JL, Stoljarova M, Warshauer DH, LaRue BL, Sajantila A, Patel J, Storts DR, Budowle B (2015) High sensitivity multiplex short tandem repeat loci analyses with massively parallel sequencing. For Sci Int Genet 16:38–47
Zeng X, King J, Hermanson S, Patel J, Storts DR, Budowle B (2015) An evaluation of the PowerSeq™ auto system: a multiplex short tandem repeat marker kit compatible with massively parallel sequencing. For Sci Int Genet 19:172–179
Fordyce SL, Avila-Arcos MC, Rockenbauer E, Borsting C, Frank-Hansen R, Petersen FT, Willerslev E, Hansen AJ, Morling N, Gilbert MT (2011) High-throughput sequencing of core STR loci for forensic genetic investigations using the Roche Genome Sequencer FLX platform. BioTechniques 51:127–133
Fordyce SL, Mogensen HS, Børsting C, Lagacé RE, Chang CW, Rajagopalan N, Morling N (2015) Second-generation sequencing of forensic STRs using the Ion Torrent™ HID STR 10-plex and the Ion PGM™. For Sci Int Genet 14:132–140
Friis SL, Buchard A, Rockenbauer E, Borsting C, Morling N (2016) Introduction of the Python script STRinNGS for analysis of STR regions in FASTQ or BAM files and expansion of the Danish STR sequence database to 11 STRs. For Sci Int Genet 21:68–75
Hoogenboom J, van der Gaag KJ, de Leeuw RH, Sijen T, de Knijff P, Laros JF (2017) FDSTools: a software package for analysis of massively parallel sequencing data with the ability to recognise and correct STR stutter and other PCR or sequencing noise. For Sci Int Genet 27:27–40
Mikkelsen M, Hansen RF, Hansen AJ, Morling N (2014) Massively parallel pyrosequencing of the mitochondrial genome with the 454 methodology in forensic genetics. For Sci Int Genet 12:30–37
King JL, Wendt FR, Sun J, Budowle B (2017) STRait Razor v2s: advancing sequence-based STR allele reporting and beyond to other marker systems. For Sci Int Genet 29:21–28
Novroski NM, King JL, Churchill JD, Seah LH, Budowle B (2016) Characterization of genetic sequence variation of 58 STR loci in four major population groups. For Sci Int Genet 25:214–226
Van Neste C, Vandewoestyne M, Van Criekinge W, Deforce D, Van Nieuwerburgh F (2014) My-Forensic-Loci-queries (MyFLq) framework for analysis of forensic STR data generated by massive parallel sequencing. For Sci Int Genet 9:1–8
Parson W, Strobl C, Huber G, Zimmermann B, Gomes SM, Souto L, Fendt L, Delport R, Langit R, Wootton S, Lagacé R, Irwin J (2013) Evaluation of next generation mtGenome sequencing using the ion torrent personal genome machine (PGM). For Sci Int Genet 7:543–549
King JL, LaRue BL, Novroski NM, Stoljarova M, Seo SB, Zeng X, Warshauer DH, Davis CP, Parson W, Sajantila A, Budowle B (2014) High quality and high-throughput massively parallel sequencing of the human mitochondrial genome using the Illumina MiSeq. For Sci Int Genet 12:128–135
Seo SB, Zeng X, King JL, Larue BL, Assidi M, Al-Qahtani MH, Sajantila A, Budowle B (2015) Underlying data for sequencing the mitochondrial genome with the massively parallel sequencing platform Ion Torrent™ PGM™. BMC Genomics 16(Suppl 1):S4
Eduardoff M, Santos C, de la Puente M, Gross M, Strobl C, Sobrino B, Ballard D, Schneider PM, Carracedo Á, Lareu MV, Parson W, Phillips C (2015) Inter-laboratory evaluation of SNP-based forensic identification by massively parallel sequencing using the Ion PGM. For Sci Int Genet 17:110–121
Churchill JD, Chang J, Ge J, Rajagopalan N, Lagacé R, Liao W, King JL, Budowle B (2015) Blind study evaluation illustrates utility of the Ion PGM™ System for use in human identity DNA typing. Croat Med J 56:218–229
Churchill JD, Schmedes SE, King JL, Budowle B (2015) Evaluation of the Illumina® beta version ForenSeq™ DNA Signature Prep Kit for use in genetic profiling. For Sci Int Genet 20:20–29
Ion AmpliSeq™ library preparation for human identification applications (2015) https://tools.thermofisher.com/content/sfs/manuals/MAN0010640_AmpliSeqLibraryPrep_HID_UG.pdf
ForenSeq™ DNA signature prep reference guide (2015) https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/forenseq/forenseq-dna-signature-prep-guide-15049528-01.pdf
Pionzio AM, McCord BR (2014) The effect of internal control sequence and length on the response to PCR inhibition in real-time PCR quantitation. For Sci Int Genet 9:55–60
Guo F, Shen H, Tian H, Jin P, Jiang X (2014) Development of a 24-locus multiplex system to incorporate the core loci in the Combined DNA Index System (CODIS) and the European Standard Set (ESS). For Sci Int Genet 8:44–54
Zhou H, Wu D, Chen R, Xu Y, Xia Z, Guo Y, Zhang F, Zheng W (2014) Developmental validation of a forensic rapid DNA-STR kit: Expressmarker 16. For Sci Int Genet 11:31–38
Core Team R (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Jäger AC, Alvarez ML, Davis CP, Guzmán E, Han Y, Way L, Walichiewicz P, Silva D, Pham N, Caves G, Bruand J, Schlesinger F, Pond SJ, Varlaro J, Stephens KM, Holt CL (2017) Developmental validation of the MiSeq FGx forensic genomics system for targeted next generation sequencing in forensic DNA casework and database laboratories. For Sci Int Genet 28:52–70
Zeng X, King JL, Budowle B (2017) Investigation of the STR loci noise distribution of PowerSeq™ auto system. Croat Med J 58:214–221
Acknowledgements
The authors would like to thank the individuals who provided the biological sample used in this study.
Funding
This project was supported in full by the National Institute of Justice (NIJ) Award Number 2015-DN-BX-K066 awarded to Sam Houston State University (SHSU) and the University of North Texas Health Science Center (UNTHSC).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplemental Table 1
(XLSX 16 kb)
Supplemental Table 2
(XLSX 14 kb)
Supplemental Table 3
(XLSX 17 kb)
Supplemental Table 4
(XLSX 17 kb)
Supplemental Table 5
(XLSX 14 kb)
Supplemental Table 6
(XLSX 14 kb)
Supplemental Table 7
(XLSX 16 kb)
Supplemental Table 8
(XLSX 14 kb)
Supplemental Table 9
(XLSX 16 kb)
Supplemental Table10
(XLSX 19 kb)
Supplemental Table 11
(XLSX 14 kb)
Supplemental Table 12
(XLSX 16 kb)
Supplemental Table 13
(XLSX 18 kb)
Supplemental Table 14
(XLSX 14 kb)
Supplemental Table 15
(XLSX 16 kb)
Supplemental Fig. 1
The average SNP loci read depth (Ion AmpliSeq™ Library Kit and ID panel) of a 1 ng DNA sample spiked with five concentrations of five PCR inhibitors. Concentration 0 means no inhibitor added. The inhibitor concentrations are listed in Table 1. Data presented as average ± standard deviation (N = 3) (XLSX 12 kb)
Supplemental Fig. 2
The average STR loci read depth (ForenSeq™ DNA Signature Prep Kit) of a 1 ng DNA sample spiked with five concentrations of five PCR inhibitors. Concentration 0 means no inhibitor added. The inhibitor concentrations were listed in Table 1. Data presented as average ± standard deviation (N = 3) (XLSX 15 kb)
Supplemental Fig. 3
The average SNP loci read depth (ForenSeq™ DNA Signature Prep Kit) of a 1 ng DNA sample spiked with five concentrations of five PCR inhibitors. Concentration 0 means no inhibitor added. The inhibitor concentrations were listed in Table 1. Data presented as average ± standard deviation (N = 3) (XLSX 15 kb)
Supplemental Fig. 4
The heatmap of alleles typed at each SNP locus with five concentrations of melanin (Ion AmpliSeq™ Library Kit and ID panel) (TIFF 60 kb)
Supplemental Fig. 5
The heatmap of alleles typed at each STR locus with five concentrations of melanin (ForenSeq™ DNA Signature Prep Kit) (TIFF 48 kb)
Supplemental Fig. 6
The heatmap of alleles typed at each SNP locus with five concentrations of melanin (ForenSeq™ DNA Signature Prep Kit) (TIFF 55 kb)
Supplemental Fig. 7
The heatmap of alleles typed at each SNP locus with five concentrations of hematin (Ion AmpliSeq™ Library Kit and ID panel) (TIFF 57 kb)
Supplemental Fig. 8
The heatmap of alleles typed at each STR locus with five concentrations of hematin (ForenSeq™ DNA Signature Prep Kit) (TIFF 35 kb)
Supplemental Fig. 9
The heatmap of alleles typed at each SNP locus with five concentrations of hematin (ForenSeq™ DNA Signature Prep Kit) (TIFF 40 kb)
Supplemental Fig. 10
The heatmap of alleles typed at each SNP locus with five concentrations of collagen (Ion AmpliSeq™ Library Kit and ID panel) (TIFF 54 kb)
Supplemental Fig. 11
The heatmap of alleles typed at each STR locus with five concentrations of collagen (ForenSeq™ DNA Signature Prep Kit) (TIFF 42 kb)
Supplemental Fig. 12
The heatmap of alleles typed at each SNP locus with five concentrations of collagen (ForenSeq™ DNA Signature Prep Kit) (TIFF 54 kb)
Supplemental Fig. 13
The heatmap of alleles typed at each SNP locus with five concentrations of calcium (Ion AmpliSeq™ Library Kit and ID panel) (TIFF 55 kb)
Supplemental Fig. 14
The heatmap of alleles typed at each STR locus with five concentrations of calcium (ForenSeq™ DNA Signature Prep Kit) (TIFF 36 kb)
Supplemental Fig. 15
The heatmap of alleles typed at each SNP locus with five concentrations of calcium (ForenSeq™ DNA Signature Prep Kit) (TIFF 41 kb)
Supplemental Fig. 16
The noise percentages of SNPs (Ion AmpliSeq™ Library Kit and ID panel) of reference samples and DNA samples spiked with five concentrations of melanin. X axis is locus read depth, Y axis is noise percentage (TIFF 153 kb)
Supplemental Fig. 17
The noise percentages of SNPs (Ion AmpliSeq™ Library Kit and ID panel) of reference samples and DNA samples spiked with five concentrations of hematin. X axis is locus read depth, Y axis is noise percentage (TIFF 132 kb)
Supplemental Fig. 18
The noise percentages of SNPs (Ion AmpliSeq™ Library Kit and ID panel) of reference samples and DNA samples spiked with five concentrations of collagen. X axis is locus read depth, Y axis is noise percentage (TIFF 154 kb)
Supplemental Fig. 19
The noise percentages of SNPs (Ion AmpliSeq™ Library Kit and ID panel) of reference samples and DNA samples spiked with five concentrations of calcium. X axis is locus read depth, Y axis is noise percentage (TIFF 153 kb)
Supplemental Fig. 20
The noise percentages of STRs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of humic acid. X axis is locus read depth, Y axis is noise percentage (TIFF 148 kb)
Supplemental Fig. 21
The noise percentages of STRs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of melanin. X axis is locus read depth, Y axis is noise percentage (TIFF 134 kb)
Supplemental Fig. 22
The noise percentages of STRs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of hematin. X axis is locus read depth, Y axis is noise percentage (TIFF 159 kb)
Supplemental Fig. 23
The noise percentages of STRs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of collagen. X axis is locus read depth, Y axis is noise percentage (TIFF 170 kb)
Supplemental Fig. 24
The noise percentages of STRs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of calcium. X axis is locus read depth, Y axis is noise percentage (TIFF 155 kb)
Supplemental Fig. 25
The noise percentages of SNPs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of humic acid. X axis is locus read depth, Y axis is noise percentage (TIFF 155 kb)
Supplemental Fig. 26
The noise percentages of SNPs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of melanin. X axis is locus read depth, Y axis is noise percentage (TIFF 136 kb)
Supplemental Fig. 27
The noise percentages of SNPs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of hematin. X axis is locus read depth, Y axis is noise percentage (TIFF 146 kb)
Supplemental Fig. 28
The noise percentages of SNPs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of collagen. X axis is locus read depth, Y axis is noise percentage (TIFF 136 kb)
Supplemental Fig. 29
The noise percentages of SNPs (ForenSeq™ DNA Signature Prep Kit) of reference samples and DNA samples spiked with five concentrations of calcium. X axis is locus read depth, Y axis is noise percentage (TIFF 132 kb)
Rights and permissions
About this article
Cite this article
Elwick, K., Zeng, X., King, J. et al. Comparative tolerance of two massively parallel sequencing systems to common PCR inhibitors. Int J Legal Med 132, 983–995 (2018). https://doi.org/10.1007/s00414-017-1693-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-017-1693-4