Abstract
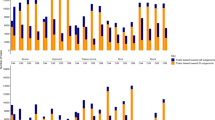
The Ph1 gene is the principal regulator of homoeologous chromosome pairing control (HECP) that ensures the diploid-like meiotic chromosome pairing behavior of polyploid wheat. The HECP control was speculated to have evolved after the first event of polyploidization. With the objective to accurately understand the evolution of the HECP control, wild emmer wheat accessions previously known to differ for HECP control were characterized for the structure and expression of the candidate Ph1 gene, C-Ph1. The C-TdPh1-5A and 5B gene copies of emmer wheat showed 98 and 99% DNA sequence similarity respectively with the corresponding hexaploid wheat copies. Further, the C-TdPh1-5B carried the C-Ph1-5B specific structural changes and transcribed three splice variants as observed in the hexaploid wheat. Further, single nucleotide changes differentiating accessions varying for HECP control were identified. Analyzed by quantitative expression analysis, the wild emmer accessions with HECP control showed ~ 10,000-fold higher transcript abundance of the C-TdPh1-5B copy during prophase-I compared to accessions lacking the control. Differential transcriptional regulation of C-TdPh1-5B splice variants further revealed that C-Ph1-5Balt1 variant is mainly responsible for differential accumulation of C-Ph1-5B copy in accessions with HECP control. Taken together, these results showed that the HECP control evolved via transcriptional regulation of splice variants during meiosis.





Similar content being viewed by others
References
Al-Kaff N, Knight E, Bertin I, Foote T, Hart N, Griffiths S, Moore G (2008) Detailed dissection of the chromosomal region containing the Ph1 locus in wheat triticum aestivum: with deletion mutants and expression profiling. Ann Bot 101:863–872
Bennypaul HS, Mutti JS, Rustgi S, Kumar N, Okubara PA, Gill KS (2012) Virus-induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Functional Integrative Genomics 12:143–156
Bhullar R, Nagarajan R, Bennypaul H, Sidhu GK, Sidhu G, Rustgi S, von Wettstein D, Gill KS (2014) Silencing of a metaphase I-specific gene results in a phenotype similar to that of the pairing homeologous 1 (Ph1) gene mutations. Proc Natl Acad Sci 111:14187–14192
Chen J, Tresenrider A, Chia M, McSwiggen DT, Spedale G, Jorgensen V, Liao H, van Werven FJ, Unal E (2017) Kinetochore inactivation by expression of a repressive mRNA. eLife 6
Chen M, Lyu G, Han M, Nie H, Shen T, Chen W, Niu Y, Song Y, Li X, Li H, Chen X, Wang Z, Xia Z, Li W, Tian XL, Ding C, Gu J, Zheng Y, Liu X, Hu J, Wei G, Tao W, Ni T (2018) 3′ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res 28:285–294
Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, Jovanovic M, Brar GA (2018) Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell 172:910–923.e16
Crismani W, Baumann U, Sutton T, Shirley N, Webster T, Spangenberg G, Langridge P, Able JA (2006) Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat. BMC Genomics 7:267
Daud HM, Gustafson JP (1996) Molecular evidence for Triticum speltoides as a B-genome progenitor of wheat (Triticum aestivum). Genome 39:543–548
Deveshwar P, Bovill WD, Sharma R, Able JA, Kapoor S (2011) Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol 11:78
Dukowic-Schulze S, Harris A, Li J, Sundararajan A, Mudge J, Retzel EF, Pawlowski WP, Chen C (2014) Comparative transcriptomics of early meiosis in Arabidopsis and maize. Journal of Genetics and Genomics 41:139–152
Dvorák J, Zhang HB (1990) Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci 87:9640–9644
Dvořák J, Terlizzi P, Zhang H, Resta P (1993) The evolution of polyploid wheats: identification of the A genome donor species. Genome 36:21–31
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Feldman M (1966) The effect of chromosomes 5B, 5D, and 5A on chromosomal pairing in Triticum aestivum. Genetics 55:1447–1453
Gill KS, Gill BS (1991) A dna fragment mapped within the submicroscopic deletion of ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 129:257–259
Gill KS, Gill BS, Endo TR, Mukai Y (1993) Fine physical mapping of Ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 134:1231–1236
Jauhar PP, Almouslem AB, Peterson TS, Joppa LR (1999) Inter- and intragenomic chromosome pairing in haploids of durum wheat. J Hered 90:437–445
Kihara H (1944) Discovery of the DD-analyser, one of the ancestors of vulgare wheats. Agric Hortic 19:889–890
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
McFadden E, Sears E (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered 37:81–89
Mello-Sampayo T. 1968. Homoeologous chromosome pairing in pentaploid hybrids of wheat. Proceedings of 3rd International Wheat Genetics Symposium: 179–184
Mello-Sampayo T (1971) Genetic regulation of meiotic chromosome pairing by chromosome 3D of Triticum aestivum. Nat New Biol 230:22–23
Mello-Sampayo T, Canas A. 1973. Suppressors of meiotic chromosome pairing in common wheat. Proceedings of 4th International Wheat Genetics Symposium: 709–713
Miller MP, Ünal E, Brar GA, Amon A (2012) Meiosis I chromosome segregation is established through regulation of microtubule–kinetochore interactions. eLife 1:e00117
Okamoto M (1957) Asynaptic effect of chromosome V. Wheat Information Service 5
Ozkan H, Feldman M (2001) Genotypic variation in tetraploid wheat affecting homoeologous pairing in hybrids with Aegilops peregrina. Genome 44:1000–1006
Randhawa HS, Mutti JS, Kidwell K, Morris CF, Chen X, Gill KS (2009) Rapid and targeted introgression of genes into popular wheat cultivars using marker-assisted background selection. PLoS One 4:e5752
Riley R, Chapman V (1958) Genetic control of the cytologically diploid behavior of hexaploid wheat. Nature 182:713–715
Riley R, Chapman V, Kimber G (1959) Genetic control of chromosome pairing in intergeneric hybrids with wheat. Nature 183:1244–1246
Riley R, Chapman V, Young RM, Belfield AM (1966) Control of meiotic chromosome pairing by the chromosomes of homoeologous group 5 of Triticum aestivum. Nature 212:1475–1477
Sarkar P, Stebbins GL (1956) Morphological evidence concerning the origin of the b genome in wheat. Am J Bot 43:297–304
Sears ER (1977) An induced mutant with homoeologous pairing in wheat. Can J Genet Cytol 19:585–593
Sears ER, Okamoto M. 1958. Intergenomic chromosome relationships in hexaploid wheat. Proceedings of 10th International Wheat Genetics Symposium 258–259
Sidhu GK, Rustgi S, Shafqat MN, von Wettstein D, Gill KS (2008) Fine structure mapping of a gene-rich region of wheat carrying Ph1, a suppressor of crossing over between homoeologous chromosomes. Proc Natl Acad Sci 105:5815–5820
Stack SM, Brown DB, Dewey WC (1977) Visualization of interphase chromosomes. J Cell Sci 26:281 LP–281299
Steber CM, Esposito RE (1995) UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression (sporulation/yeast/transcription/URSI promoter element). Genetics 92:12490–12494
Tyankova DN (2001) Euploid and monosomic intergeneric wheat x Aegilops ovata hybrids: i. production and cytological characteristics. Comptes Rendus de l’Academie Bulgare des Sciences 54:5:77–5:77
Zickler D, Kleckner N (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7:a016626
Acknowledgments
We would like to thank Dr. Moshe Feldman for kindly sharing the germplasm.
Author’s contribution
KS and KSG designed the research. KS and MAK performed the experiments and data analysis. KS and KSG wrote the manuscript. KS, MAK, and KSG critically revised the manuscript.
Funding
This work was supported by USDA National Institute of Food and Agriculture (Hatch project number WNP00449) and United States Agency for International Development Feed the Future Innovation Lab-Climate Resilient Wheat (Grant number AID-OAA-A-13-00008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Summary:
Ph1 locus plays a very critical role in chromosome pairing and recombination in polyploid wheat. The Ph1 locus prohibits recombination between chromosomes of polyploid wheat and its wild relatives leading to restricted or no crossing over. Thus, making it difficult to transfer gene(s) of interest from wild relatives to cultivated wheat. Until recently, very limited information is available about the gene controlling the phenotype. Recent identification of candidate Ph1 (C-Ph1) paved the way for further understanding its evolution, and mechanism of action would pave the way for its targeted use in wheat crop improvement. In this study, we have identified the major transcript regulating C-Ph1 gene expression that can be used in future studies for targeted manipulation of recombination to transfer biotic and abiotic stress tolerance from wild relatives of polyploid wheat.
Electronic supplementary material
Fig. S1
Multiple sequence alignment of C-TdPh1 and C-TaPh1 gene homoeologs around major copy specific structural changes (29 bp deletion and 60 bp insertion) highlighting conservation of gene structure in hexaploid and tetraploid wheat. (PNG 292 kb)
Fig. S2
Multiple sequence alignment of C-TdPh1-5B gene sequence from wild emmer wheat accessions varying for homoeologous chromosome pairing control for the region carrying 29 bp deletion and 60 bp insertion. (PNG 48 kb)
Fig. S3
Multiple sequence alignment of C-Ph1-5A copy from wild emmer wheat accessions varying for HECP control. Sequence conservation at a site is highlighted with asterisk (*) at the bottom, whereas sequence change between wild emmer wheat and hexaploid wheat is highlighted with black rectangular box and asterisk (*) on top of the box. Single nucleotide variants differentiating –HECP and + HECP lines are highlighted with blue rectangular boxes and asterisk (*) on the top of the box. (PNG 275 kb)
Fig. S4
Multiple sequence alignment of C-Ph1-5B copy from wild emmer wheat accessions varying for HECP control. Sequence conservation at a site is highlighted with asterisk (*) at the bottom, whereas sequence change between wild emmer wheat and hexaploid wheat is highlighted with black rectangular box and asterisk (*) on top of the box. Single nucleotide variants differentiating –HECP and + HECP lines are highlighted with blue rectangular boxes and asterisk (*) on the top of the box. (PNG 357 kb)
Supplementary Table 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Rawale, K.S., Khan, M.A. & Gill, K.S. The novel function of the Ph1 gene to differentiate homologs from homoeologs evolved in Triticum turgidum ssp. dicoccoides via a dramatic meiosis-specific increase in the expression of the 5B copy of the C-Ph1 gene. Chromosoma 128, 561–570 (2019). https://doi.org/10.1007/s00412-019-00724-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00724-6