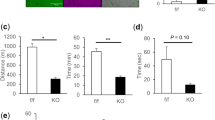
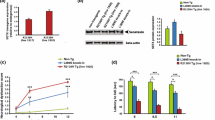
Abstract
Recently, we provided genetic basis showing that mitochondrial dysfunction can trigger motor neuron degeneration, through identification of CHCHD10 encoding a mitochondrial protein. We reported patients, carrying the p.Ser59Leu heterozygous mutation in CHCHD10, from a large family with a mitochondrial myopathy associated with motor neuron disease (MND). Rapidly, our group and others reported CHCHD10 mutations in amyotrophic lateral sclerosis (ALS), frontotemporal dementia-ALS and other neurodegenerative diseases. Here, we generated knock-in (KI) mice, carrying the p.Ser59Leu mutation, that mimic the mitochondrial myopathy with mtDNA instability displayed by the patients from our original family. Before 14 months of age, all KI mice developed a fatal mitochondrial cardiomyopathy associated with enhanced mitophagy. CHCHD10S59L/+ mice also displayed neuromuscular junction (NMJ) and motor neuron degeneration with hyper-fragmentation of the motor end plate and moderate but significant motor neuron loss in lumbar spinal cord at the end stage of the disease. At this stage, we observed TDP-43 cytoplasmic aggregates in spinal neurons. We also showed that motor neurons differentiated from human iPSC carrying the p.Ser59Leu mutation were much more sensitive to Staurosporine or glutamate-induced caspase activation than control cells. These data confirm that mitochondrial deficiency associated with CHCHD10 mutations can be at the origin of MND. CHCHD10 is highly expressed in the NMJ post-synaptic part. Importantly, the fragmentation of the motor end plate was associated with abnormal CHCHD10 expression that was also observed closed to NMJs which were morphologically normal. Furthermore, we found OXPHOS deficiency in muscle of CHCHD10S59L/+ mice at 3 months of age in the absence of neuron loss in spinal cord. Our data show that the pathological effects of the p.Ser59Leu mutation target muscle prior to NMJ and motor neurons. They likely lead to OXPHOS deficiency, loss of cristae junctions and destabilization of internal membrane structure within mitochondria at motor end plate of NMJ, impairing neurotransmission. These data are in favor with a key role for muscle in MND associated with CHCHD10 mutations.








Similar content being viewed by others
References
Auranen M, Ylikallio E, Shcherbii M, Paetau A, Kiuru-Enari S, Toppila JP et al (2015) CHCHD10 variant p.(Gly66Val) causes axonal Charcot–Marie–Tooth disease. Neurol Genet 1:e1. https://doi.org/10.1212/nxg.0000000000000003
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2014) A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain J Neurol 137:2329–2345. https://doi.org/10.1093/brain/awu138
Bannwarth S, Berg-Alonso L, Augé G, Fragaki K, Kolesar JE, Lespinasse F et al (2016) Inactivation of Pif1 helicase causes a mitochondrial myopathy in mice. Mitochondrion 30:126–137. https://doi.org/10.1016/j.mito.2016.02.005
Blasco H, Mavel S, Corcia P, Gordon PH (2014) The glutamate hypothesis in ALS: pathophysiology and drug development. Curr Med Chem 21:3551–3575
Bloch-Gallego E (2015) Mechanisms controlling neuromuscular junction stability. Cell Mol Life Sci 72:1029–1043. https://doi.org/10.1007/s00018-014-1768-z
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Cozzolino M, Rossi S, Mirra A, Carrì MT (2015) Mitochondrial dynamism and the pathogenesis of amyotrophic lateral sclerosis. Front Cell Neurosci 9:31. https://doi.org/10.3389/fncel.2015.00031
Dadon-Nachum M, Melamed E, Offen D (2011) The “Dying-Back” phenomenon of motor neurons in ALS. J Mol Neurosci 43:470–477. https://doi.org/10.1007/s12031-010-9467-1
Desport JC, Preux PM, Magy L, Boirie Y, Vallat JM, Beaufrère B et al (2001) Factors correlated with hypermetabolism in patients with amyotrophic lateral sclerosis. Am J Clin Nutr 74:328–334. https://doi.org/10.1093/ajcn/74.3.328
Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Bonconpagni S et al (2008) Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab 8:425–436. https://doi.org/10.1016/j.cmet.2008.09.002
Dupuis L, Gonzalez de Aguilar J-L, Echaniz-Laguna A, Eschbach J, Rene F, Oudart H et al (2009) Muscle mitochondrial uncoupling dismantles neuromuscular junction and triggers distal degeneration of motor neurons. PLoS One 4:e5390. https://doi.org/10.1371/journal.pone.0005390
Dupuis L, Gonzalez de Aguilar J-L, Oudart H, de Tapia M, Barbeito L, Loeffler J-P (2004) Mitochondria in amyotrophic lateral sclerosis: a trigger and a target. Neurodegener Dis 1:245–254. https://doi.org/10.1159/000085063
Dupuis L, Pradat P-F, Ludolph AC, Loeffler J-P (2011) Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol 10:75–82. https://doi.org/10.1016/S1474-4422(10)70224-6
Genin EC, Bannwarth S, Lespinasse F, Ortega-Vila B, Fragaki K, Itoh K et al (2018) Loss of MICOS complex integrity and mitochondrial damage, but not TDP-43 mitochondrial localisation, are likely associated with severity of CHCHD10-related diseases. Neurobiol Dis 119:159–171. https://doi.org/10.1016/j.nbd.2018.07.027
Genin EC, Plutino M, Bannwarth S, Villa E, Cisneros-Barroso E, Roy M et al (2016) CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med 8:58–72. https://doi.org/10.15252/emmm.201505496
Gorman GS, McFarland R, Stewart J, Feeney C, Turnbull DM (2018) Mitochondrial donation: from test tube to clinic. Lancet 392:1191–1192. https://doi.org/10.1016/S0140-6736(18)31868-3
Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R et al (2017) HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. https://doi.org/10.1038/s41467-017-00911-y
Hell K (2008) The Erv1–Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta BBA Mol Cell Res 1783:601–609. https://doi.org/10.1016/j.bbamcr.2007.12.005
Huang ML-H, Sivagurunathan S, Ting S, Jansson PJ, Austin CJD, Kelly M et al (2013) Molecular and functional alterations in a mouse cardiac model of friedreich ataxia. Am J Pathol 183:745–757. https://doi.org/10.1016/j.ajpath.2013.05.032
Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, Drory VE et al (2014) Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain J Neurol 137:e311. https://doi.org/10.1093/brain/awu265
Jones RA, Reich CD, Dissanayake KN, Kristmundsdottir F, Findlater GS, Ribchester RR et al (2016) NMJ-morph reveals principal components of synaptic morphology influencing structure–function relationships at the neuromuscular junction. Open Biol 6:160240. https://doi.org/10.1098/rsob.160240
Lehmer C, Schludi MH, Ransom L, Greiling J, Junghänel M, Exner N et al (2018) A novel CHCHD10 mutation implicates a Mia40-dependent mitochondrial import deficit in ALS. EMBO Mol Med 10:e8558. https://doi.org/10.15252/emmm.201708558
Limongelli G, Tome-Esteban M, Dejthevaporn C, Rahman S, Hanna MG, Elliott PM (2010) Prevalence and natural history of heart disease in adults with primary mitochondrial respiratory chain disease. Eur J Heart Fail 12:114–121. https://doi.org/10.1093/eurjhf/hfp186
Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M et al (2015) Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol 33:89–96. https://doi.org/10.1038/nbt.3049
Mesci P, Zaïdi S, Lobsiger CS, Millecamps S, Escartin C, Seilhean D et al (2015) System xC—is a mediator of microglial function and its deletion slows symptoms in amyotrophic lateral sclerosis mice. Brain 138:53–68. https://doi.org/10.1093/brain/awu312
Müller K, Andersen PM, Hübers A, Marroquin N, Volk AE, Danzer KM et al (2014) Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain J Neurol 137:e309. https://doi.org/10.1093/brain/awu227
Musarò A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M et al (2001) Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet 27:195–200. https://doi.org/10.1038/84839
Palma E, Inghilleri M, Conti L, Deflorio C, Frasca V, Manteca A et al (2011) Physiological characterization of human muscle acetylcholine receptors from ALS patients. Proc Natl Acad Sci 108:20184–20188. https://doi.org/10.1073/pnas.1117975108
Palma E, Reyes-Ruiz JM, Lopergolo D, Roseti C, Bertollini C, Ruffolo G et al (2016) Acetylcholine receptors from human muscle as pharmacological targets for ALS therapy. Proc Natl Acad Sci 113:3060–3065. https://doi.org/10.1073/pnas.1600251113
Penttilä S, Jokela M, Bouquin H, Saukkonen AM, Toivanen J, Udd B (2015) Late onset spinal motor neuronopathy is caused by mutation in CHCHD10. Ann Neurol 77:163–172. https://doi.org/10.1002/ana.24319
Philips T, Rothstein JD (2015) Rodent models of amyotrophic lateral sclerosis: rodent models of ALS. Curr Protoc Pharmacol 69:5.67.1–5.67.21. https://doi.org/10.1002/0471141755.ph0567s69
Raff MC (2002) Axonal self-destruction and neurodegeneration. Science 296:868–871. https://doi.org/10.1126/science.1068613
Rocha MC, Pousinha PA, Correia AM, Sebastião AM, Ribeiro JA (2013) Early Changes of neuromuscular transmission in the SOD1(G93A) mice model of ALS start long before motor symptoms onset. PLoS One 8:e73846. https://doi.org/10.1371/journal.pone.0073846
Ronchi D, Riboldi G, Del Bo R, Ticozzi N, Scarlato M, Galimberti D et al (2015) CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain J Neurol 138:e372. https://doi.org/10.1093/brain/awu384
Rustin P, Chretien D, Bourgeron T, Gérard B, Rötig A, Saudubray JM et al (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta Int J Clin Chem 228:35–51
Salmon M, Esiri M, Ruderman N (1971) Myopathic disorder associated with mitochondrial abnormalities, hyperglycaemia, and hyperketonaemia. Lancet 298:290–293. https://doi.org/10.1016/S0140-6736(71)91335-3
Smith EF, Shaw PJ, De Vos KJ (2017) The role of mitochondria in amyotrophic lateral sclerosis. Neurosci Lett. https://doi.org/10.1016/j.neulet.2017.06.052
The International Stem Cell Banking Initiative (2009) Consensus guidance for banking and supply of human embryonic stem cell lines for research purposes. Stem Cell Rev Rep 5:301–314. https://doi.org/10.1007/s12015-009-9085-x
Tong M, Sadoshima J (2016) Mitochondrial autophagy in cardiomyopathy. Curr Opin Genet Dev 38:8–15. https://doi.org/10.1016/j.gde.2016.02.006
Van Den Bosch L, Van Damme P, Bogaert E, Robberecht W (2006) The role of excitotoxicity in the pathogenesis of amyotrophic lateral sclerosis. Biochim Biophys Acta BBA Mol Basis Dis 1762:1068–1082. https://doi.org/10.1016/j.bbadis.2006.05.002
Villa E, Proïcs E, Rubio-Patiño C, Obba S, Zunino B, Bossowski JP et al (2017) Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep 20:2846–2859. https://doi.org/10.1016/j.celrep.2017.08.087
Willadt S, Nash M, Slater CR (2016) Age-related fragmentation of the motor endplate is not associated with impaired neuromuscular transmission in the mouse diaphragm. Sci Rep. https://doi.org/10.1038/srep24849
Wishart TM, Parson SH, Gillingwater TH (2006) Synaptic vulnerability in neurodegenerative disease. J Neuropathol Exp Neurol 65:733–739. https://doi.org/10.1097/01.jnen.0000228202.35163.c4
Woo J-AA, Liu T, Trotter C, Fang CC, De Narvaez E, LePochat P et al (2017) Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat Commun 8:15558. https://doi.org/10.1038/ncomms15558
Yaplito-Lee J, Weintraub R, Jamsen K, Chow CW, Thorburn DR, Boneh A (2007) Cardiac manifestations in oxidative phosphorylation disorders of childhood. J Pediatr 150:407–411. https://doi.org/10.1016/j.jpeds.2006.12.047
Yuan X, Xiao Y-C, Zhang G-P, Hou N, Wu X-Q, Chen W-L et al (2016) Chloroquine improves left ventricle diastolic function in streptozotocin-induced diabetic mice. Drug Des Dev Ther 10:2729–2737. https://doi.org/10.2147/DDDT.S111253
Acknowledgements
We thank our colleagues from Genetic Engineering and Mouse Transgenesis department at service unit CIPHE (Centre d’Immunophénomique, Aix Marseille Université, INSERM, CNRS, Phenomin, Marseille, France) and mainly Frederic Fiore who provided expertise and know-how and perform the generation of the CHCHD10S59L/+ mice that greatly assisted our project. We also thank our colleagues, including Benoit Petit-Demoulière, Ghina Bou-About, Hamid Meziane, Hugues Jacobs, Marie-France Champy, Patrick Reilly, Tania Sorg and Yann Herault from Phenomin (Institut Clinique de la souris, 1 rue Laurent Fries, 67404 ILLKIRCH cedex 2—CNRS, UMR7104, Illkirch, France—INSERM, U964, Illkirch, France—Université de Strasbourg, France) for their help in phenotyping mice. We gratefully acknowledge the IRCAN’s Molecular and Cellular Core Imaging (PICMI) facility, supported financially by FEDER, Conseil régional Provence Alpes-Côte d’Azur, Conseil Départemental 06, Cancéropôle PACA, Gis Ibisa and Inserm, the IRCAN’s Animal core facility, supported by Région Provence Alpes-Côte d’Azur and Inserm, the IRCAN’s Histology core facility, supported by Région Provence Alpes-Côte d’Azur and Université de Nice Sophia-Antipolis, the Centre Méditerranéen de Médecine Moléculaire (C3 M) imaging facility, the University’s CCMA Electron Microscopy facility supported by Université de Nice Sophia-Antipolis, Région Provence Alpes-Côte d’Azur, Conseil Départemental 06, and Gis Ibisa and, the ICM CELIS-iPS internal platform. We also thank Dr Luisa Villa and Dr Pamela Moceri for their help in interpreting muscle and cardiac results, respectively, and Sandra Foustoul and Alyssia Mari for technical help.
Funding
This work was made possible by Grants to VP-F from the ANR (Agence Nationale de la Recherche) ANR-16-CE16-0024-01, from the AFM-Téléthon (Association Française contre les Myopathies) #20947 and from the Fondation Maladies Rares. The research leading to the iPSC results has received funding from the program “Investissements d’avenir” ANR-11-INBS-0011-NeurATRIS: Translational Research Infrastructure for Biotherapies in Neurosciences”. ECG and BMH are postdoctoral fellows supported by ANR-16-CE16-0024-01. CL-O is a Ph.D. fellow supported by ANR-10-LABX-73.
Author information
Authors and Affiliations
Contributions
ECG and SBa designed, performed experiments and analyzed data on mouse models, and contributed to the main text. BMH designed, performed experiments and analyzed data on cell models (NSC34 and iPSCs-derived motor neurons), and contributed to the main text. KF designed, performed experiments and analyzed data on respiratory chain experiments. SL-G performed electron microscopy experiments. JN, AM-C, FL, BR, GA and CC performed and analyzed experiments on mouse models. SBi generated iPSC. CL-O and DB provided the motor neuron differentiation protocol and advices on the iPSC daily culture and motor neuron differentiation. DB provides advices and helped in data interpretation on iPSC-derived motor neuron experiments and contributed to the main text. AC provided technical advices for motor neurons staining and count. FM helped with the interpretation of metabolic data. SBo and CL provided advices for the NMJ and motor neurons analyses and helped in data interpretation. J‐ER designed and analyzed data on cell models and contributed to the main text. VP‐F designed, supervised the project and drafted the manuscript. All the authors read and approved the final submission.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (MP4 21757 kb)
Supplementary material 2 (MP4 50009 kb)
Supplementary Figure 1. CHCHD10 and TOM20 expression around NMJs in the gastrocnemius muscle from
+/+and S59L/+ mice. a-b. CHCHD10 and TOM20 (translocase of outer mitochondrial membrane 20) expression around NMJs in the gastrocnemius muscle from one +/+ (a) and one S59L/+ (b) mice. A maximum intensity projection of Z-stacked confocal images showed immunofluorescent staining of TOM20 (cyan), CHCHD10 (red) and α-Bungarotoxin (BTX, green) to mark NMJ (n = 2 +/+ and 2 S59L/+ mice). Scale bar: 10µm. The low panel of each image is a magnification (area indicated by the square in the upper panel) of muscle fibers around NMJ with a higher intensity of fluorescence. Scale bare: 5µm. (PPTX 1072 kb)
Supplementary Figure 2. Ultrastructural analysis of motor neurons
in the ventral horn of lumbar spinal cord from +/+ (left panels) and S59L/+ (right panels) mice showing vacuolated mitochondria with abnormal cristae in mutant animals at end stage (a). The low panel of each image is a magnification of the area indicated by the square in the middle panel. Scale bar: 10 µm (upper panels), 1 µm (middle panels) and 0.25 µm (lower panels). b. Quantification of mitochondrial morphology observed in a. Intermediary refers to mitochondria that cannot be clearly classified as abnormal despite the presence of slight cristae anomalies. Shown are mean ± SEM (n = 2 +/+ and 2 S59L/+ mice with 45 mitochondria per animal randomly chosen and analyzed). Statistical analysis was performed using Student’s t-test. P-value = *<0.05, **<0.01. (PPTX 3572 kb)
Supplementary Figure 3. Sensory–motor functions of
CHCHD10S59L/+mice. Motor abilities assessed by rotarod (a), grip (b), string (c) and crenelated beam (d) tests. EMG measurements (e-g). All experiments were performed in 10 CHCHD10S59L/+ and 10 CHCHD10+/+ mice of each sex. Data are expressed as mean ± SEM and analyzed using Student t-test. (PPTX 107 kb)
Supplementary Figure 4. Apoptosis analysis in MEFs and mice at end point, and characterization of iPSCs. a.
MEFs were isolated from two +/+ or two S59L/+ mice (S59L-1 and -2) and treated with 1 µM STS for 8 hours and 16 hours (O/N). Cell death was determined by flow cytometry using Annexin V/DAPI staining. n=3 per condition. b-c. DEVDase activity measurement (b) and Western-Blot analysis of pro-caspase 3 (C3) and cleaved caspase-3 (CC3) (c) on brain, spinal cord, muscle and heart protein extracts. d-g. Characterization of iPSC clones. d. Representative immunofluorescent staining of the S59L/+ iPSC clone for pluripotency markers TRA1-60, NANOG and SSEA-4, and the differentiation marker SSEA-1. Nuclei are stained with Hoechst. Scale bars: 100 µm. e. Representative immunofluorescent staining of plated EBs for lineage-specific markers: nestin for ectoderm, SMA for mesoderm, and β-catenin for endoderm. Scale bars: 100 µm. f. Pluripotency and tri-lineage differentiation potential were characterized by TaqMan® hPSC Scorecard™. Results are expressed as a score relative to a reference set of standard iPSC and ESC clones. g. Karyogram show genome integrity of the S59L/+ iPSC clone compared to parental fibroblasts. Illumina SNP array data were analyzed using Karyostudio. An autosomal detected region deviating from reference data is annotated with orange band (deletion). This alteration was below the level that would be detected by G-banding and was present in both parental fibroblasts and the derived iPSC clone. h. Phase contrast images of iPSC-derived motor neuron from control (C1, C2), and S59L/+ lines at day 35 (scale bar, 100 µm). Differences between groups were analyzed by two-way ANOVA (Tukey’s multiple comparisons test). P-value = *<0.05, **<0.01. (PPTX 1271 kb)
Supplementary Figure 5. Heart analysis of
CHCHD10S59L/+mice at 3 months of age. a. Spectrophotometric analysis of respiratory chain in heart from +/+ and S59L/+ mice. Results represent the mean ± SD of 3 +/+ and 3 S59L/+ mice. b. Representative western blot analysis from equal amounts (2.5μg) of heart homogenates of +/+ and S59L/+ mice using OXPHOS antibodies cocktail detecting complexes I to V and anti-GAPDH antibody for loading control. c. Quantification of relative intensities of OXPHOS proteins shown in b. Data are shown as the mean ± SD of 3 independent experiments (n= 3 +/+ and 3 S59L/+ mice). d. Long extension PCR of mtDNA from heart of 6 +/+ (lanes 3-8) and 6 S59L/+ (lanes 9-14) mice. mtDNA indicates the 12.8kb amplicon. Lanes 1,2: molecular weight markers. Lane 13: negative PCR control. e. Mitochondrial DNA quantification in heart of +/+ or S59L/+ mice. The mouse mitochondrial 12S rRNA (mtDNA) and the nuclear GAPDH (nDNA) genes were individually amplified by real-time PCR. Data were expressed as ratio between mtDNA and nDNA concentration. Results represent the mean of relative PCR ± SD of 3 independent experiments from 4 +/+ and 5 S59L/+ mice. The data from S59L/+ mice were compared to the controls. Statistical analysis was performed using Student’s t-test in a and Mann-Whitney’s test in c and e. P-value = **<0.01, ***<0.001. (PPTX 9061 kb)
Rights and permissions
About this article
Cite this article
Genin, E.C., Madji Hounoum, B., Bannwarth, S. et al. Mitochondrial defect in muscle precedes neuromuscular junction degeneration and motor neuron death in CHCHD10S59L/+ mouse. Acta Neuropathol 138, 123–145 (2019). https://doi.org/10.1007/s00401-019-01988-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-01988-z