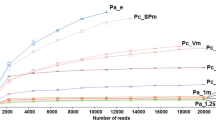
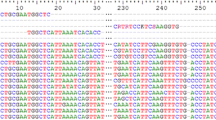
Abstract
Peritrich ciliates are highly diverse and can be important bacterial grazers in aquatic ecosystems. Morphological identifications of peritrich species and assemblages in the environment are time-consuming and expertise-demanding. In this study, two peritrich-specific PCR primers were newly designed to amplify a fragment including the internal transcribed spacer (ITS) region of ribosomal rDNA from environmental samples. The primers showed high specificity in silico, and in tests with peritrich isolates and environmental DNA. Application of these primers in clone library construction and sequencing yielded exclusively sequences of peritrichs for water and sediment samples. We also found the ITS1, ITS2, ITS, D1 region of 28S rDNA, and ITS+D1 region co-varied with, and generally more variable than, the V9 region of 18S rDNA in peritrichs. The newly designed specific primers thus provide additional tools to study the molecular diversity, community composition, and phylogeography of these ecologically important protists in different systems.
Similar content being viewed by others
References
Altschul S F, Madden T L, Schäffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. 1997. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Research, 25 (17): 3 389–3 402, https://doi.org/10.1093/nar/25.17.3389.
Amaral-Zettler L A, McCliment E A, Ducklow H W, Huse S M. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One, 4 (7): e6372, https://doi.org/10.1371/journal.pone.0006372.
Bachy C, Dolan J R, López-García P, Deschamps P, Moreira D. 2013. Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. The ISME Journal, 7 (2): 244–255, https://doi.org/10.1038/ismej.2012.106.
Clamp J C, Coats D W. 2000. Planeticovorticella finleyi n.g., n.sp. (Peritrichia, Vorticellidae), a planktonic ciliate with a polymorphic life cycle. Invertebrate Biology, 119 (1): 1–16, https://doi.org/10.1111/j.1744-7410.2000.tb00169.x.
Foissner W, Agatha S, Berger H. 2002. Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Biologiezentrum des Oberösterreichischen Landesmuseums, Linz, Austria. 1 459p.
Foissner W, Berger H, Kohmann F. 1991. Taxonomische und okologische Revision der Ciliaten des Saprobiensystems. Band II: Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft, 5/92: 1–502.
Fried J, Mayr G, Berger H, Traunspurger W, Psenner R, Lemmer H. 2000. Monitoring protozoa and metazoa biofilm communities for assessing wastewater quality impact and reactor up-scaling effects. Water Science and Technology, 41 (4–5): 309–316.
Fu R, Gong J. 2017. Single cell analysis linking ribosomal (r) DNA and rRNA copy numbers to cell size and growth rate provides insights into molecular protistan ecology. Journal of Eukaryotic Microbiology, https://doi.org/10.1111/jeu.12425.
Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics, 12 (6): 543–548, https://doi.org/10.1093/bioinformatics/12.6.543.
Gentekaki E, Lynn D. 2012. Spatial genetic variation, phylogeography and barcoding of the peritrichous ciliate Carchesium polypinum. European Journal of Protistology, 48 (4): 305–313, https://doi.org/10.1016/j.ejop.2012.04.001.
Gong J, Dong J, Liu X H, Massana R. 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist, 164 (3): 369–379, https://doi.org/10.1016/j.protis.2012.11.006.
Guo X H, Zhang Q Q, Zhang X L, Zhang J S, Gong J. 2015. Marine fungal communities in water and surface sediment of a sea cucumber farming system: habitat-differentiated distribution and nutrients driving succession. Fungal Ecology, 14: 14–87. https://doi.org/10.1016/j.funeco.2014.12.001.
Hall T A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41 (2): 95–98.
Hillis D M, Dixon M T. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology, 66 (4): 411–453.
Huang J R, Lin W H, Zeng W, Xu R L. 2005. The effect of sediment restoration on the protozoan community in shrimp culture ponds. Ecologic Science, 24 (4): 326–329. (in Chinese with English abstract)
Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics, 20 (14): 2 317–2 319, https://doi.org/10.1093/bioinformatics/bth226.
Ji D D, Song W B, Al-Rasheid K A S, Li L F. 2005. Taxonomic characterization of two marine peritrichous ciliates, Pseudovorticella clampi n. sp. and Zoothamnium pararbuscula n. sp. (Ciliophora: Peritrichia), from North China. Journal of Eukaryotic Microbiology, 52 (2): 159–169, https://doi.org/10.1111/j.1550-7408.2005.05-3353.x.
Ji D D, Sun P, Warren A, Song W B. 2009. Colonial sessilid peritrichs. In: Song W B, Warren A, Hu X eds. Free-living ciliates in the Bohai and Yellow Seas, China. Science Press, Beijing, China. p.257-286. (in Chinese)
Katoh K, Misawa K, Kuma K I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30 (14): 3 059–3 066, https://doi.org/10.1093/nar/gkf436.
Liu X H, Gong J. 2012. Revealing the diversity and quantity of Peritrich ciliates in environmental samples using specific primer-based PCR and quantitative PCR. Microbes and Environments, 27 (4): 497–503, https://doi.org/10.1264/jsme2.ME12056.
Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environmental Microbiology, 10 (10): 2 894–2 898, https://doi.org/10.1111/j.1462-2920.2008.01706.x.
Markmann M, Tautz D. 2005. Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philosophical Transactions of the Royal Society B: Biological Sciences, 360 (1462): 1 917–1 924, https://doi.org/10.1098/rstb.2005.1723.
Martín-Cereceda M, Serrano S, Guinea A. 2001. Biofilm communities and operational monitoring of a rotating biological contactor system. Water, Air, and Soil Pollution, 126 (3–4): 193–206, https://doi.org/10.1023/A:1005291015122.
Maurin L C, Himmel D, Mansot J L, Gros O. 2010. Raman microspectrometry as a powerful tool for a quick screening of thiotrophy: an application on mangrove swamp meiofauna of Guadeloupe (F.W.I.). Marine Environmental Research, 69 (5): 382–389, https://doi.org/10.1016/j.marenvres.2010.02.001.
Orsi W, Biddle J F, Edgcomb V. 2013. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One, 8 (2): e56335, https://doi.org/10.1371/journal.pone.0056335.
Rocap G, Distel D L, Waterbury J B, Chisholm S W. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Applied and Environmental Microbiology, 68 (3): 1 180–1 191, https://doi.org/10.1128/AEM.68.3.1180-1191.2002.
Ronquist F, Huelsenbeck J P. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics, 19 (12): 1 572–1 574, https://doi.org/10.1093/bioinformatics/btg180.
Schloss P D, Westcott S L, Ryabin T, Hall J R, Hartmann M, Hollister E B, Lesniewski R A, Oakley B B, Parks D H, Robinson C J, Sahl J W, Stres B, Thallinger G G, Van Horn D J, Weber C F. 2009. Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75 (23): 7 537–7 541, https://doi.org/10.1128/AEM.01541-09.
Schoch C L, Seifert K A, Huhndorf S, Robert V, Spouge J L, Levesque C A, Chen W, Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 109 (16): 6 241–6 246, https://doi.org/10.1073/pnas.1117018109.
Sherr B F, Sherr E B, Rassoulzadegan F. 1988. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature dependence. Applied and Environmental Microbiology, 54 (5): 1 091–1 095.
Shi X L, Liu X J, Liu G J, Sun Z Q, Xu H L. 2012. An approach to analyzing spatial patterns of protozoan communities for assessing water quality in the Hangzhou section of Jing-Hang Grand Canal in China. Environmental Science and Pollution Research, 19 (3): 739–747, https://doi.org/10.1007/s11356-011-0615-0.
Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30 (9): 1 312–1 313, https://doi.org/10.1093/bioinformatics/btu033.
Stern R F, Andersen R A, Jameson I, Küpper F C, Coffroth M A, Vaulot D, Le Gall F, Véron B, Brand J J, Skelton H, Kasai F, Lilly E L, Keeling P J. 2012. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One, 7 (8): e42780, https://doi.org/10.1371/journal.pone.0042780.
Sun P, Clamp J C, Xu D P, Huang B Q, Shin M K, Turner F. 2013. An ITS-based phylogenetic framework for the genus Vorticella: finding the molecular and morphological gaps in a taxonomically difficult group. Proceedings of the Royal Society B: Biological Science, 280 (1771): 20131177, https://doi.org/10.1098/rspb.2013.1177.
Sun P, Clamp J, Xu D P, Kusuoka Y, Miao W. 2012. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade: redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist, 163 (1): 129–142, https://doi.org/10.1016/j.protis.2011.06.005.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30 (12): 2 725–2 729, https://doi.org/10.1093/molbev/mst197.
Xu H L, Min G S, Choi J K, Jung J H, Park M H. 2009. An approach to analyses of periphytic ciliate colonization for monitoring water quality using a modified artificial substrate in Korean coastal waters. Marine Pollution Bulletin, 58 (9): 1 278–1 285, https://doi.org/10.1016/j.marpolbul.2009.05.003.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (Nos. 31572255, 41522604, 31301867), the Strategic Priority Research Program of CAS (No. XDA11020702), and the Science and Technology Development Program of Yantai (No. 2014ZH073)
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Su, L., Zhang, Q. & Gong, J. Development and evaluation of specific PCR primers targeting the ribosomal DNA-internal transcribed spacer (ITS) region of peritrich ciliates in environmental samples. J. Ocean. Limnol. 36, 818–826 (2018). https://doi.org/10.1007/s00343-018-6326-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-018-6326-3