Abstract
Key message
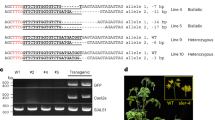
Base editing in tomatoes was achieved by transient expression.
Abstract
The Solanaceae plants, particularly the tomato (Solanum lycopersicum), is of huge economic value worldwide. The tomato is a unique model plant for studying the functions of genes related to fruit ripening. Deeper understanding of tomatoes is of great importance for both plant research and the economy. Genome editing technology, such as CRISPR/Cas9, has been used for functional genetic research. However, some challenges, such as low transformation efficiency, remain with this technology. Moreover, the foreign Cas9 and gRNA expression cassettes must be removed to obtain null-segregants In this study, we used a high-level transient expression system to improve the base editing technology. A high-level transient expression system has been established previously using geminiviral replication and a double terminator. The pBYR2HS vector was used for this transient expression system. nCas9-CDA and sgRNA-SlHWS were introduced into this vector, and the protein and RNA were then transiently expressed in tomato tissues by agroinfiltration. The homozygous mutant produced by base editing was obtained in the next generation with an efficiency of about 18%. nCas9-free next-generation plants were 71%. All the homozygous base-edited plants in next generation are nCas9-free. These findings show that the high-level transient expression system is useful for base editing in tomatoes.





Similar content being viewed by others
Abbreviations
- AID:
-
Activation-induced cytidine deaminase
- ALS:
-
Acetolactate synthase
- BeYDV:
-
Bean yellow dwarf virus
- Cas9:
-
CRISPR-associated endonuclease 9
- CDA:
-
Cytidine deaminase
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- DSB:
-
Double strand break
- GABA:
-
γ-Aminobutyric acid
- Indel:
-
Insertion and/or deletion
- Rep:
-
Replication
- LIR:
-
Long intergenic region
- sgRNA:
-
Single guide RNA
- SlHWS:
-
Solanum lycopersicum HAWAIIAN SKIRT
References
Aliaga-Franco N, Zhang C, Presa S, Srivastava AK, Granell A, Alabadi D, Sadanandom A, Blazquez MA, Minguet EG (2019) Identification of transgene-free CRISPR-edited plants of rice, tomato, and Arabidopsis by monitoring DsRED fluorescence in dry seeds. Front Plant Sci 10:1150
Bánfalvi Z, Csákvári E, Villányi V, Kondrák M (2020) Generation of transgene-free PDS mutants in potato by Agrobacterium-mediated transformation. BMC Biotechnol 20:25
Bransteitter R, Pham P, Scharff MD, Goodman MF (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A 100:4102–4107
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Chaudhary P, Sharma A, Singh B, Nagpal AK (2018) Bioactivities of phytochemicals present in tomato. J Food Sci Technol 55:2833–2849
Chen L, Li W, Katin-Grazzini L, Ding J, Gu X, Li Y, Gu T, Wang R, Lin X, Deng Z, McAvoy RJ, Gmitter FG Jr, Deng Z, Zhao Y, Li Y (2018) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res 5:13
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Damayanti F, Lombardo F, Masuda JI, Shinozaki Y, Ichino T, Hoshikawa K, Okabe Y, Wang N, Fukuda N, Ariizumi T, Ezura H (2019) Functional disruption of the tomato putative ortholog of HAWAIIAN SKIRT results in facultative parthenocarpy, reduced fertility and leaf morphological defects. Front Plant Sci 10:1234
Demirer GS, Zhang H, Matos JL, Goh NS, Cunningham FJ, Sung Y, Chang R, Aditham AJ, Chio L, Cho MJ, Staskawicz B, Landry MP (2019) High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol 14:456–464
Endo M, Mikami M, Endo A, Kaya H, Itoh T, Nishimasu H, Nureki O, Toki S (2019) Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat Plants 5:14–17
Gao X, Chen J, Dai X, Zhang D, Zhao Y (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171:1794–1800
Gonzalez-Carranza ZH, Zhang X, Peters JL, Boltz V, Szecsi J, Bendahmane M, Roberts JA (2017) HAWAIIAN SKIRT controls size and floral organ number by modulating CUC1 and CUC2 expression. PLoS ONE 12:e0185106
Hahn F, Eisenhut M, Mantegazza O, Weber APM (2018) Homology-directed repair of a defective glabrous gene in Arabidopsis with Cas9-based gene targeting. Front Plant Sci 9:424
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788
Hoshikawa K, Fujita S, Renhu N, Ezura K, Yamamoto T, Nonaka S, Ezura H, Miura K (2019) Efficient transient protein expression in tomato cultivars and wild species using agroinfiltration-mediated high expression system. Plant Cell Rep 38:75–84
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467:76–82
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, Kotake-Nara E, Kawasaki S, Toki S (2017) Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat Plants 3:866–874
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Kitajima S, Miura K, Yasuda J (2020) Radish sprouts as an efficient and rapidly available host for an agroinfiltration-based transient gene expression system. Plant Biotechnol 37(1):89–92
Knapp S, Bohs L, Nee M, Spooner DM (2004) Solanaceae a model for linking genomics with biodiversity. Comp Funct Genom 5:285–291
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Lai H, He J, Engle M, Diamond MS, Chen Q (2012) Robust production of virus-like particles and monoclonal antibodies with geminiviral replicon vectors in lettuce. Plant Biotechnol J 10:95–104
Li Q, Sapkota M, van der Knaap E (2020) Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: unlocking the neglected potential for crop improvement. Hortic Res 7:36
Liu Y, Zeng J, Yuan C, Guo Y, Yu H, Li Y, Huang C (2019) Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol J 17:1191–1193
Luo Z, Chen Z (2007) Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19:943–958
Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38:84–89
Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y (2005) Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 23:718–723
Miura K, Sato A, Shiba H, Kang SW, Kamada H, Ezura H (2012) Accumulation of antioxidants and antioxidant activity in tomato, Solanum lycopersicum, are enhanced by the transcription factor SlICE1. Plant Biotechnol 29:261–269
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353:aaf8729
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361:1259–1262
Nonaka S, Someya T, Zhou S, Takayama M, Nakamura K, Ezura H (2017) An Agrobacterium tumefaciens strain with gamma-aminobutyric acid transaminase activity shows an enhanced genetic transformation ability in plants. Sci Rep 7:42649
Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP (2014) Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53
Quinet M, Angosto T, Yuste-Lisbona FJ, Blanchard-Gros R, Bigot S, Martinez JP, Lutts S (2019) Tomato fruit development and metabolism. Front Plant Sci 10:1554
Rizvi I, Choudhury NR, Tuteja N (2015) Insights into the functional characteristics of geminivirus rolling-circle replication initiator protein and its interaction with host factors affecting viral DNA replication. Arch Virol 160:375–387
Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52:283–296
Sato S, Tabata S, Hirakawa H et al (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Suzaki T, Tsuda M, Ezura H, Day B, Miura K (2019) Agroinfiltration-based efficient transient protein expression in leguminous plants. Plant Biotechnol 36:119–123
Veillet F, Perrot L, Chauvin L, Kermarrec M-P, Guyon-Debast A, Chauvin J-E, Nogué F, Mazier M (2019) Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci 20:402
Yamamoto T, Hoshikawa K, Ezura K, Okazawa R, Fujita S, Takaoka M, Mason HS, Ezura H, Miura K (2018) Improvement of the transient expression system for production of recombinant proteins in plants. Sci Rep 8:4755
Zhang X, Jayaweera D, Peters JL, Szecsi J, Bendahmane M, Roberts JA, Gonzalez-Carranza ZH (2017) The Arabidopsis thaliana F-box gene HAWAIIAN SKIRT is a new player in the microRNA pathway. PLoS ONE 12:e0189788
Acknowledgements
We thank Ms. Kazuko Ito and Ms. Yuri Nemoto at Tsukuba-Plant Innovation Research Center (T-PIRC), University of Tsukuba for technical support. Tomato seeds (cv. Micro-Tom, TOMJPF00001) were provided from T-PIRC, University of Tsukuba through the National Bio-Resource Project (NBRP) of the Japan Agency for Research and Development (AMED), Japan. This work was supported by the Cross-ministerial Strategic Innovation Promotion Program (SIP), Japan, Program on Open Innovation Platform with Enterprises, Research Institute and Academia, Japan Science and Technology Agency (JST, OPERA, JPMJOP1851), and a Cooperative Research Grant from the Plant Transgenic Design Initiative, Gene Research Center, T-PIRC, University of Tsukuba.
Author information
Authors and Affiliations
Contributions
TA, HE, and KM conceived and designed the experiments; KN and AK provided materials and contributed to interpretation of data. SY, IMYA, and SK conducted the experiments and collected data. SY, IMYA, TA, and KM wrote the initial draft of the manuscript. All authors critically reviewed the manuscript and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Additional information
Communicated by Hiroyasu Ebinuma.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, S., Kawasaki, S., Abdellatif, I.M.Y. et al. Efficient base editing in tomato using a highly expressed transient system. Plant Cell Rep 40, 667–676 (2021). https://doi.org/10.1007/s00299-021-02662-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02662-z