Abstract
Key message
Several hypomethylated sites within the Karma region of EgDEF1 and hotspot regions in chromosomes 1, 2, 3, and 5 may be associated with mantling.
Abstract
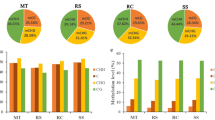
One of the main challenges faced by the oil palm industry is fruit abnormalities, such as the “mantled” phenotype that can lead to reduced yields. This clonal abnormality is an epigenetic phenomenon and has been linked to the hypomethylation of a transposable element within the EgDEF1 gene. To understand the epigenome changes in clones, methylomes of clonal oil palms were compared to methylomes of seedling-derived oil palms. Whole-genome bisulfite sequencing data from seedlings, normal, and mantled clones were analyzed to determine and compare the context-specific DNA methylomes. In seedlings, coding and regulatory regions are generally hypomethylated while introns and repeats are extensively methylated. Genes with a low number of guanines and cytosines in the third position of codons (GC3-poor genes) were increasingly methylated towards their 3′ region, while GC3-rich genes remain demethylated, similar to patterns in other eukaryotic species. Predicted promoter regions were generally hypomethylated in seedlings. In clones, CG, CHG, and CHH methylation levels generally decreased in functionally important regions, such as promoters, 5′ UTRs, and coding regions. Although random regions were found to be hypomethylated in clonal genomes, hypomethylation of certain hotspot regions may be associated with the clonal mantling phenotype. Our findings, therefore, suggest other hypomethylated CHG sites within the Karma of EgDEF1 and hypomethylated hotspot regions in chromosomes 1, 2, 3 and 5, are associated with mantling.










Similar content being viewed by others
Abbreviations
- WGBS:
-
Whole-genome bisulfite sequencing
- TFBS:
-
Transcription factor binding site
- TE:
-
Transposable element
- mCG:
-
Methylation at CG context
- mCHG:
-
Methylation at CHG context
- mCHH:
-
Methylation at CHH context
References
Anastasiadi D, Esteve-Codina A, Piferrer F (2018) Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 11:37. https://doi.org/10.1186/s13072-018-0205-1
An Y-QC, Goettel W, Han Q et al (2017) Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci Rep 7:12263. https://doi.org/10.1038/s41598-017-12510-4
Bartels A, Han Q, Nair P et al (2018) Dynamic DNA methylation in plant growth and development. Int J Mol Sci 19:2144. https://doi.org/10.3390/ijms19072144
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Cantu D, Vanzetti LS, Sumner A et al (2010) Small RNAs, DNA methylation and transposable elements in wheat. BMC Genomics 11:408. https://doi.org/10.1186/1471-2164-11-408
Chan K-L, Rosli R, Tatarinova TV et al (2017a) Seqping: gene prediction pipeline for plant genomes using self-training gene models and transcriptomic data. BMC Bioinformatics 18:1426. https://doi.org/10.1186/s12859-016-1426-6
Chan K-L, Tatarinova TV, Rosli R et al (2017b) Evidence-based gene models for structural and functional annotations of the oil palm genome. Biol Direct 12:21. https://doi.org/10.1186/s13062-017-0191-4
Dapp M, Reinders J, Bédiée A et al (2015) Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants 1:15092. https://doi.org/10.1038/nplants.2015.92
Deng X, Song X, Wei L et al (2016) Epigenetic regulation and epigenomic landscape in rice. Natl Sci Rev 3:309–327. https://doi.org/10.1093/nsr/nww042
Elhamamsy AR (2016) DNA methylation dynamics in plants and mammals: overview of regulation and dysregulation. Cell Biochem Funct 34:289–298. https://doi.org/10.1002/cbf.3183
Francescatto M, Pardo L, Rizzu P et al (2011) Profiling transcription initiation in human aged brain using deep-CAGE. BMC Bioinformatics 12:A8. https://doi.org/10.1186/1471-2105-12-S11-A8
Fujimoto R, Uezono K, Ishikura S et al (2018) Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed Sci 68:145–158. https://doi.org/10.1270/jsbbs.17155
Gallusci P, Dai Z, Génard M et al (2017) Epigenetics for plant improvement: current knowledge and modeling avenues. Trends Plant Sci 22:610–623. https://doi.org/10.1016/j.tplants.2017.04.009
Ghosh J, Coutifaris C, Sapienza C, Mainigi M (2017) Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin Epigenetics 9:14. https://doi.org/10.1186/s13148-017-0318-6
Han Z, Crisp PA, Stelpflug S et al (2018) Heritable epigenomic changes to the maize methylome resulting from tissue culture. Genetics 209:983–995. https://doi.org/10.1534/genetics.118.300987
Hashim AT, Ishak Z, Rosli SK et al (2018) Oil palm (Elaeis guineensis Jacq.) somatic embryogenesis. In: Jain SM, Gupta P (eds) Step wise protocols for somatic embryogenesis of important woody plants. Springer International Publishing, Berlin, pp 209–229
Hossain MS, Kawakatsu T, Kim KD et al (2017) Divergent cytosine DNA methylation patterns in single-cell, soybean root hairs. New Phytol 214:808–819. https://doi.org/10.1111/nph.14421
Hsieh T-F, Ibarra CA, Silva P et al (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454. https://doi.org/10.1126/science.1172417
Hsu F-M, Gohain M, Allishe A et al (2018) Dynamics of the methylome and transcriptome during the regeneration of rice. Epigenomes 2:14. https://doi.org/10.3390/epigenomes2030014
Kel AE, Gössling E, Reuter I et al (2003) MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31:3576–3579. https://doi.org/10.1093/nar/gkg585
Kim D, Pertea G, Trapnell C et al (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. https://doi.org/10.1186/gb-2013-14-4-r36
Komatsu M, Shimamoto K, Kyozuka J (2003) Two-Step Regulation and Continuous Retrotransposition of the Rice LINE-Type Retrotransposon Karma. Plant Cell 15:1934–1944. https://doi.org/10.1105/tpc.011809
Kooke R, Johannes F, Wardenaar R et al (2015) Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 27:337–348. https://doi.org/10.1105/tpc.114.133025
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. https://doi.org/10.1093/bioinformatics/btr167
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Latzel V, Allan E, Bortolini Silveira A et al (2013) Epigenetic diversity increases the productivity and stability of plant populations. Nat Commun 4:2875. https://doi.org/10.1038/ncomms3875
Lauss K, Wardenaar R, Oka R et al (2018) Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol 176:1627–1645. https://doi.org/10.1104/pp.17.01054
Li G, Jia Q, Zhao J et al (2014) Dysregulation of genome-wide gene expression and DNA methylation in abnormal cloned piglets. BMC Genomics 15:811. https://doi.org/10.1186/1471-2164-15-811
Li H, Handsaker B, Wysoker A et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lin J-Y, Le BH, Chen M et al (2017) Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. PNAS 114:E9730–E9739. https://doi.org/10.1073/pnas.1716758114
Lioznova AV, Khamis AM, Artemov AV et al (2019) CpG traffic lights are markers of regulatory regions in human genome. BMC Genomics. https://doi.org/10.1186/s12864-018-5387-1
Liu H, Li S, Wang X et al (2016) DNA methylation dynamics: identification and functional annotation. Brief Funct Genomics 15:470–484. https://doi.org/10.1093/bfgp/elw029
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Martin A, Troadec C, Boualem A et al (2009) A transposon-induced epigenetic change leads to sex determination in melon. Nature 461:1135–1138. https://doi.org/10.1038/nature08498
Medvedeva YA, Khamis AM, Kulakovskiy IV et al (2014) Effects of cytosine methylation on transcription factor binding sites. BMC Genomics 15:119. https://doi.org/10.1186/1471-2164-15-119
Michael TP (2014) Plant genome size variation: bloating and purging DNA. Brief Funct Genomics 13:308–317. https://doi.org/10.1093/bfgp/elu005
Murphy DJ (2007) Future prospects for oil palm in the 21st century: Biological and related challenges. Eur J Lipid Sci Technol 109:296–306. https://doi.org/10.1002/ejlt.200600229
Nagai M, Meguro-Horike M, Horike SI (2012) Epigenetic defects related reproductive technologies: large offspring syndrome (LOS), DNA Methylation - From Genomics to Technology, Tatiana Tatarinova and Owain Kerton, IntechOpen. https://doi.org/10.5772/34102
Niederhuth CE, Bewick AJ, Ji L et al (2016) Widespread natural variation of DNA methylation within angiosperms. Genome Biol 17:194. https://doi.org/10.1186/s13059-016-1059-0
Niederhuth CE, Schmitz RJ (2017) Putting DNA methylation in context: from genomes to gene expression in plants. Biochim Biophys Acta Gene Regul Mech 1860:149–156. https://doi.org/10.1016/j.bbagrm.2016.08.009
Novakovic B, Lewis S, Halliday J et al (2019) Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nat Commun 10:3922. https://doi.org/10.1038/s41467-019-11929-9
Ong-Abdullah M, Ordway JM, Jiang N et al (2015) Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525:533–537. https://doi.org/10.1038/nature15365
Pardo LM, Rizzu P, Francescatto M et al (2013) Regional differences in gene expression and promoter usage in aged human brains. Neurobiol Aging 34:1825–1836. https://doi.org/10.1016/j.neurobiolaging.2013.01.005
Piegu B, Guyot R, Picault N et al (2006) Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269. https://doi.org/10.1101/gr.5290206
Pikaard CS, Mittelsten Scheid O (2014) Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 6:a019315. https://doi.org/10.1101/cshperspect.a019315
Rishi V, Bhattacharya P, Chatterjee R et al (2010) CpG methylation of half-CRE sequences creates C/EBP binding sites that activate some tissue-specific genes. Proc Natl Acad Sci 107:20311–20316. https://doi.org/10.1073/pnas.1008688107
Sablok G, Nayak KC, Vazquez F, Tatarinova TV (2011) Synonymous codon usage, GC3, and evolutionary patterns across plastomes of three pooid model species: emerging grass genome models for monocots. Mol Biotechnol 49:116–128. https://doi.org/10.1007/s12033-011-9383-9
Sanusi NSNM, Nik Nik Shazana, Rosli R et al (2018) PalmXplore: oil palm gene database. Database. https://doi.org/10.1093/database/bay095
Singh R, Ong-Abdullah M, Low E-TL et al (2013) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500:335–339. https://doi.org/10.1038/nature12309
Siretskiy A, Spjuth O (2014) HTSeq-Hadoop: Extending HTSeq for massively parallel sequencing data analysis using Hadoop. In: 2014 IEEE 10th International Conference on e-Science. pp 317–323
Soh AC, Wong G, Tan CC et al (2011) Commercial-scale propagation and planting of elite oil palm clones: research and development towards realization. J Oil Palm Res 23:935–952
Song Q-X, Lu X, Li Q-T et al (2013) Genome-wide analysis of DNA methylation in soybean. Mol Plant 6:1961–1974. https://doi.org/10.1093/mp/sst123
Stelpflug SC, Eichten SR, Hermanson PJ et al (2014) Consistent and heritable alterations of DNA methylation are induced by tissue culture in maize. Genetics 198:209–218. https://doi.org/10.1534/genetics.114.165480
Stroud H, Ding B, Simon SA et al (2013) Plants regenerated from tissue culture contain stable epigenome changes in rice. Elife 2:e00354. https://doi.org/10.7554/eLife.00354
Tanurdzic M, Vaughn MW, Jiang H et al (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6:e302. https://doi.org/10.1371/journal.pbio.0060302
Tatarinova T, Elhaik E, Pellegrini M (2013) Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol Evol 5:1443–1456. https://doi.org/10.1093/gbe/evt103
Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA (2010) GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics 11:308. https://doi.org/10.1186/1471-2164-11-308
Tatarinova TV, Chekalin E, Nikolsky Y et al (2016) Nucleotide diversity analysis highlights functionally important genomic regions. Sci Rep 6:35730. https://doi.org/10.1038/srep35730
Triska M, Grocutt D, Southern J et al (2013) cisExpress: motif detection in DNA sequences. Bioinformatics 29:2203–2205. https://doi.org/10.1093/bioinformatics/btt366
Triska M, Solovyev V, Baranova A et al (2017) Nucleotide patterns aiding in prediction of eukaryotic promoters. PLoS ONE 12:e0187243. https://doi.org/10.1371/journal.pone.0187243
Troukhan M, Tatarinova T, Bouck J et al (2009) Genome-wide discovery of cis-elements in promoter sequences using gene expression. OMICS 13:139–151. https://doi.org/10.1089/omi.2008.0034
Wang P, Xia H, Zhang Y et al (2015) Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may). BMC Genomics 16:21. https://doi.org/10.1186/s12864-014-1204-7
Wen G (2017) A simple process of RNA-Sequence analyses by Hisat2, Htseq and DESeq2. In: Proceedings of the 2017 International Conference on Biomedical Engineering and Bioinformatics - ICBEB 2017. ACM Press, New York, New York, USA, pp 11–15
West PT, Li Q, Ji L et al (2014) Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS ONE 9:e105267. https://doi.org/10.1371/journal.pone.0105267
Wingender E, Kel AE, Kel OV et al (1997) TRANSFAC, TRRD and COMPEL: towards a federated database system on transcriptional regulation. Nucleic Acids Res 25:265–268. https://doi.org/10.1093/nar/25.1.265
Yang H, Chang F, You C et al (2015) Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J 81:268–281. https://doi.org/10.1111/tpj.12726
Yan H, Bombarely A, Xu B et al (2018) siRNAs regulate DNA methylation and interfere with gene and lncRNA expression in the heterozygous polyploid switchgrass. Biotechnol Biofuels 11:208. https://doi.org/10.1186/s13068-018-1202-0
Zemach A, Kim MY, Silva P et al (2010) Local DNA hypomethylation activates genes in rice endosperm. PNAS 107:18729–18734. https://doi.org/10.1073/pnas.1009695107
Zhang D, Wang Z, Wang N et al (2014) Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS ONE 9:e96879. https://doi.org/10.1371/journal.pone.0096879
Zhang Y-Y, Fischer M, Colot V, Bossdorf O (2013) Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 197:314–322. https://doi.org/10.1111/nph.12010
Acknowledgements
We would like to thank the Director-General of MPOB for permission to publish this study. Our appreciation goes to the Bioinformatics Unit, Breeding and Tissue Culture Unit of MPOB, especially Epigenetics Group and MPOB Bagan Datuk Research Station, Malaysia for their technical support throughout this study. We thank the University of La Verne for the support of this international collaboration.
Funding
The research was supported by the MPOB.
Author information
Authors and Affiliations
Contributions
S-EO and NS performed the experiment, TT, AE, and ET analyzed the data, NS, S-EO, TT, E-TLL, and MO-A interpreted the findings. TT, E-TLL, and MO-A supervised the work. All authors participated in the preparation of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
None to declare.
Consent for publication
All authors agreed to the publication and approved the final version.
Availability of data and material
The central oil palm database is https://palmxplore.mpob.gov.my/palmXplore/. Whole-genome bisulfite sequence data can be assessed in the NCBI SRA under accession numbers SAMN03569063—SAMN03569077. The cDNA sequence of the kDEF1 transcript is available in GenBank under accession number KR347486. RNA-sequencing data can be accessed under GEO Accession No. GSE144079.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sarpan, N., Taranenko, E., Ooi, SE. et al. DNA methylation changes in clonally propagated oil palm. Plant Cell Rep 39, 1219–1233 (2020). https://doi.org/10.1007/s00299-020-02561-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02561-9