Abstract
Key message
We summarize recent progress of CRISPR/Cas9-mediated gene targeting in plants, provide recommendations for designing gene-targeting vectors and highlight the potential of new technologies applicable to plants.
Abstract
Gene targeting (GT) is a tool of urgent need for plant biotechnology and breeding. It is based on homologous recombination that is able to precisely introduce desired modifications within a target locus. However, its low efficiency in higher plants is a major barrier for its application. Using site-specific nucleases, such as the recent CRISPR/Cas system, GT has become applicable in plants, via the induction of double-strand breaks, although still at a too low efficiency for most practical applications in crops. Recently, a variety of promising new improvements regarding the efficiency of GT has been reported by several groups. It turns out that GT can be enhanced by cell-type-specific expression of Cas nucleases, by the use of self-amplified GT-vector DNA or by manipulation of DNA repair pathways. Here, we highlight the most recent progress of GT in plants. Moreover, we provide suggestions on how to use the technology efficiently, based on the mechanisms of DNA repair, and highlight several of the newest GT strategies in yeast or mammals that are potentially applicable to plants. Using the full potential of GT technology will definitely help us pave the way in enhancing crop yields and food safety for an ecologically friendly agriculture.
Similar content being viewed by others
What is gene targeting?
Gene targeting (GT) is a technology utilized to enable genome modifications through homologous recombination (HR) (Paszkowski et al. 1988). HR is an endogenous mechanism that functions to restore damaged DNA or to trigger crossovers between homologues during meiosis. However, HR activity is low in higher plants. As a result, GT techniques are rarely applied—neither in basic scientific research nor in biotechnology. This is mainly due to the fact that higher plants predominantly use the non-homologous end joining (NHEJ) pathway for DNA repair in somatic cells (Puchta 2005). NHEJ is an error-prone DNA repair mechanism that tends to cause mutations which are not predicable on the sequence level. It was shown some time ago that site-specific nucleases are able to induce double-strand breaks (DSBs) at particular loci and stimulate both NHEJ (Salomon and Puchta 1998) and HR-based GT (Puchta et al. 1996). Site-specific nucleases based on the CRISPR/Cas system are the most important development in recent biotechnology for genome engineering due to their ease in application (Schindele et al. 2018). Several studies demonstrate the application of site-specific nuclease-enhanced GT in many important crops, such as rice, maize and wheat (Endo et al. 2016; Gil-Humanes et al. 2017; Li et al. 2018a, b; Sun et al. 2016a; Svitashev et al. 2015, 2016; Wang et al. 2017). This review will highlight recent progress of GT in plants and combine it with previous knowledge of HR to provide recommendations for GT designs.
Cellular components influence gene targeting
Over the years, various approaches have been used to enhance GT efficiencies in plants, that have been extensively reviewed (Puchta and Fauser 2013; Steinert et al. 2016; Sun et al. 2016b). Here, we focus on the most recent developments in manipulating DNA repair pathways for the improvement of GT techniques in the future.
Across several kingdoms, a number of studies demonstrated that GT or HR efficiency can be increased by blocking the NHEJ pathway or enhancing the HR pathway. In Arabidopsis, mutants of ku70 and lig4 were reported to have higher GT efficiencies, with up to a 19-fold increase (Qi et al. 2013). Similarly, using CRISPR/Cas9 to knockout Ligase 4 altogether with GT approach achieved enhanced GT in rice (Endo et al. 2016). As an alternative strategy, it was shown that overexpression of some heterologous proteins involved in HR repair enhanced GT effectively, such as overexpressing the RAD54 protein of yeast in plants (Shaked et al. 2005). Overexpressing Arabidopsis RAD54 specifically in egg cells increased GT in Arabidopsis by tenfold (Even-Faitelson et al. 2011). Interestingly, expression of the bacterial RecA stimulated intrachromosomal HR but not GT (Reiss et al. 1996, 2000).
On the other hand, recent experiments in mammalian cells indicate that the blocking of NHEJ might not always lead to enhanced GT events. Polymerase Q, a main factor of microhomology-mediated NHEJ, also mediates T-DNA integration in Arabidopsis (van Kregten et al. 2016). In mammalian systems, a cell line in which both ligase 4 and polymerase Q were knocked out, did not show an increase of GT events but showed no random integration of GT-vectors in comparison with the wild-type cell lines (Saito et al. 2017; Zelensky et al. 2017). These experiments, although no DSB was induced in the target locus, nevertheless, indicate that depending on the GT efficiency calculating method, the ratio of GT to random integration events can be increased by blocking NHEJ-mediated events without a real enhancement of HR-mediated events.
There is another downside to when DNA repair mutants are used or HR proteins are over-expressed: genomic instabilities will be induced in these cells and the respective transgenes or mutations have to be removed via additional steps such as backcrossing, thus limiting the usefulness of these methods. One might consider transient approaches such as gene silencing to suppress genes without causing permanent modifications of the genome. Nowadays, the developing toolbox of CRISPR/Cas provides more possibilities to suppress NHEJ pathways and promote HR pathways: a dead Cas9 fusion with a transcription suppressor or activator can be used to suppress or enhance mRNA expression, respectively, via binding to the promoter region (La Russa and Qi 2015). CRISPR/Cas13, a type of RNA-guided RNase, is able to post-transcriptionally control gene expression (Abudayyeh et al. 2017); however, it has not been utilized to facilitate GT in plants thus far (for review see Wolter and Puchta 2018). If we are able to successfully integrate previous knowledge of DNA repair pathways with GT, these advancing techniques will prove to be powerful tools for genome engineering in the future.
Evidence has accumulated that GT efficiency is influenced by the cell cycle. In the G1 phase of yeast and human cells, the NHEJ pathway is the dominant repair system for DSBs, whilst HR repair is more active in the S and G2 phases (Ferreira and Cooper 2004; Mao et al. 2008). Moreover, it has been demonstrated that GT efficiencies are higher in the G2/M cell cycle phase in human pluripotent stem cells, and in the S-phase of Saccharomyces cerevisiae and several other fungi (Tsakraklides et al. 2015; Yang et al. 2016). The influence of the cell cycle on GT efficiency might be caused by various factors, from changes in chromatin structure to activation of DNA repair during replication. The accessibility of DNA can have a dramatic influence on GT, which was demonstrated following a mutation in the chromatin assembly factor FAS1, resulting in increased HR frequencies (Endo et al. 2006; Kirik et al. 2006). Similarly, GT efficiency is also influenced by cell type. Taking mouse as an example, their embryonic stem cells exhibit substantial GT, with efficiencies up to 100% (Miura et al. 2018; Quadros et al. 2017); however, GT efficiency in somatic cells still remains low. Although the influence of cell cycle and cell types affects GT, the detailed roles of these factors on GT remain unclear in plant cells. Recent reports revealed that the egg cell of Arabidopsis could be an excellent cell type to perform GT, which could achieve promising frequencies of inheritable GT events (Miki et al. 2018; Wolter et al. 2018).
Recommended design for CRISPR/Cas9 target site mutation
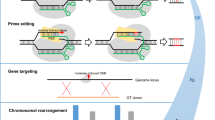
Schematically, GT-mediated genome modifications can be classified into three categories—insertions, replacements and substitutions, although classification can be fluent (Fig. 1a). Insertions add additional sequences into the genome. An example of this is the introduction of GFP tags into open reading frames used for tracking proteins microscopically. Replacements remove long stretches of genomic sequences and replace them with desired sequences, for example, a weak wild-type promoter can be replaced by a stronger or a constitutive one. The last kind of change, substitutions, introduces only minor changes into the genome, such as single-base exchanges that result in amino acid substitutions. Since the rapidly advancing quantitative trait locus (QTL) mapping techniques are extensively used to identify QTLs of many beneficial traits, using GT can help introduce these QTLs, especially SNPs, into various important agricultural cultivars of the same species, saving the time required for crossing in traditional breeding practices. In addition, some combinations of beneficial traits might not be achievable via crossing due to their genetic position on allelic chromosomes. Moreover, it is worth to mention that the CRISPR/Cas-derived base-editor technology, through deamination of cytidines or adenines, is able to achieve single nucleotide substitutions within the genome of crops (Kim et al. 2017; Li et al. 2017, 2018b; Zong et al. 2017), which could be a valuable alternative of GT.
Recommended design for CRISPR/Cas9 target site mutation. a The three major categories of GT-mediated genome modifications—insertion, replacements, substitutions. b–d To prevent GT-vector DNA and final GT products being cut by Cas9, the Cas9 targeting site has to be eliminated either by insertion (b), modification of the PAM sequence (c) or by changes in the protospacer-binding sequence (d). The mechanism of GT according to the SDSA. Using the design in c as an example, only one strand directly induced by Cas9 has the perfect homology for SDSA pathway, but another strand does not (e–g). The ideal strand has a perfect homologous region for strand invasion and will end up with the desired modification in the target site (e). This non-ideal strand initiates strand invasion within its homology, but will not end up with a modification in the target locus (f). If DNA degradation occurs naturally, and the homologous region is exposed, SDSA pathway-mediated GT is still possible (g). Dash-line: newly synthesized DNA
When it comes to the detailed planning of GT experiments, one has to take a number of problems into consideration. While sequence-specific DSBs are the most effective method to increase GT efficiencies, an issue arises to prevent the GT-vector and GT-modified genome from destruction by unintended DSB induction. Using GT experiments with an insertion, for example, we depict several possibilities when Streptococcus pyogenes Cas9 (SpCas9) is used in Fig. 1. First, if the insertion site is positioned next to the Cas9 targeting site, including the protospacer adjacent motif (PAM) region which is NGG for SpCas9, the Cas9 target site is simply eliminated due to the inserted sequence (Fig. 1b). Alternatively, if the SpCas9 targeted region and PAM region is not overlapping with the inserted sequence, one has to modify the PAM sequence (e.g. NGG to NCG) in the GT-vector (Fig. 1c). From PAM sequence activity assays of SpCas9, we know most changes from a G to an A, or a T to a C are sufficient to remove cleavage activity, but one should avoid the use of NAG because this is an active but weaker PAM sequence for SpCas9 (Hsu et al. 2013). It is also recommended to apply Staphylococcus aureus Cas9 (SaCas9) in GT. In Arabidopsis, SaCas9 displays more efficient mutagenesis via DSB-based gene editing (Fauser et al. 2014; Steinert et al. 2015). Similarly, SaCas9-mediated GT showed higher GT efficiency than with the application of SpCas9 (Schiml et al. 2014; Wolter et al. 2018). These results are in line with a recent biochemical study showing that SaCas9 has a much higher reaction turnover than SpCas9 (Yourik et al. 2019). Following analysis in plants, it was reported that the targeting efficiency of SaCas9 is much lower with non-canonical PAMs (Kaya et al. 2016); therefore, modifying PAM sequences other than the canonical PAM (NNGRRT) is feasible to hinder SaCas9 targeting within the GT-vector. However, the Cas9-targeted region can also be removed via modification of the protospacer recognizing region complementary to the single-stranded guide RNA, by the introduction of several nucleotide changes (Fig. 1d). In general, modification of the PAM region is the most effective way to eliminate DNA cleavage activity of Cas9. However, we recommend to construct the GT-vector sequence in such a way that all newly introduced mutations are on only one end of the DSB site in the target locus (Fig. 1c, d). This recommendation is based on our understanding of GT and HR mechanisms in plants. It had been demonstrated that during GT using T-DNAs as the template, most DSBs are repaired by the SDSA pathway in plant somatic cells (Puchta 1998). The same mechanisms also operate when an intrachromosomal homology is used for repair (Orel et al. 2003). This synthesis-based DNA repair consists of 5′ end resection, homology searching and 3′ end invasion for priming and elongation of the repair template (Fig. 1e, g). When the DSB is induced by Cas9, the two ends of the DSB are resected and become single-stranded DNAs (ssDNA) with free 3′ ends. A 3′ end which is homologous to the GT-vector sequence will displace one strand of the GT-vector DNA, forming the displacement loop (D-loop) (Fig. 1e, f). This process is called 3′ end invasion. Afterwards, this invading 3′ ssDNA is used as a primer for elongation, replicating the sequence information from the GT-vector DNA. Subsequently, the new synthesized ssDNA dissociates from the D-loop. As this strand now contains homology to the other DSB end, reannealing and bridging the break is thus possible. Therefore, following further elongation and ligation, a modified target incorporating the sequence information of the GT-vector is obtained. This mechanism is different from classical crossover events between homologous chromosomes during meiosis, as no resolution of recombination intermediates by nucleases is required (Puchta 2005). However, 3′ end DNA might be trimmed by additional degradation of DNA (Fig. 1g). Depending on which 3′ end is invading, HR could merely lead to the repair of the break (Fig. 1f) or successful GT (Fig. 1e, g).
What would happen if we design modified sequences at both flanking sites corresponding to the DSB? In Fig. 2a, we depicted this scenario showing two potential outcomes (Fig. 2b, c). First of all, both resected 3′ ssDNA ends would not have perfect homology to the GT-vector. These heterologies would reduce the efficiency of both homology searching and D-loop formation. Even if 3′ end invasion occurs correctly and the sequence information is successfully copied to the target site, either the original PAM sequence may be retained (Fig. 2b) or the insertion would not be copied from the GT-vector to the target site (Fig. 2c). Without modification of the PAM sequence within the target, the Cas9 would be able to generate a DSB at this GT event, risking additional mutagenesis within the target. Furthermore, another possibility is that only the modified PAM would be copied to the target site. Ultimately, the desired insertion would not be incorporated into the target genomic locus. Therefore, we do not recommend designing the PAM and insertion mutations in the GT-vector on opposite ends relative to the DSB site of the target. However, a GT event which includes both modifications is possible: the initial free 3′ DNA generated via Cas9 cleavage might also be degraded or resected like a 5′ DNA strand, similar to the degradation shown in Fig. 1g. This resected 3′ single-stranded DNA can use the SDSA pathway to incorporate all sequence modifications. Currently, the influence of heterologies in GT-vector remains elusive in plants, leaving this question open for future experimentation.
Heterologous sequences at opposite positions of DSB cause failed GT. a The desired insertion and PAM sequence modifications are designed at opposite sites in the GT-vector, marked in red. b The mismatch at the 3′ end will reduce the efficiency of homology search and 3′ end invasion. Even if a successful invasion occurred, the modified PAM will not be incorporated into the target, leading to further mutations due to DSB induction within the target locus. c Alternatively, the 3′ DNA end has the perfect homology for invasion, although this event will only lead to the incorporation of the mutation at the PAM site but not the desired insertion. Dash-line: newly synthesized DNA. (Color figure online)
Aberrant GT due to a combination of HR and NHEJ
One has to keep in mind that due to the fact that the SDSA is the prevailing mechanism of homologous DSB repair in somatic cells, even if a reaction is initiated by the invasion of a homologous 3′ end, as depicted in Fig. 1, there is no necessity that HR occurs at the other end of the DSB, too. Using a GT-mediated insertion as an example (Fig. 3a), if the correct ssDNA is used for 3′ end invasion (Fig. 3b) and followed up by a template switch event, this two-side GT event creates the ideal modification in the target locus (Fig. 3c). Alternatively, second end capture might also be achieved by NHEJ with unpredictable outcomes in terms of modifications (Fig. 3d). Early on in DSB-induced GT experiments in plants, besides perfect events by HR, events were also found in which one junction was repaired by HR and the other by NHEJ (Puchta et al. 1996). Using T-DNAs that contained homology to only one end of the break actually enabled integration with almost the same efficiency as T-DNAs that carried homologies to both ends (Puchta 1998). Another class of events has also been found that are due to a combination of HR and NHEJ, as it is also possible that the 3′ end of the vector DNA is invading the target site (Fig. 3e). Thus, homologies are copied from the target to the vector at one or the other of its ends. In a second independent step, this vector integrates elsewhere at an ectopic site within the genome, by NHEJ. This process is also referred to as ectopic targeting (e.g. Hanin et al. 2001). Therefore, it is definitely required to investigate the modified loci after GT in detail, e.g. sequencing the complete modified locus, to prove that the both newly formed junctions arose via HR at the target locus.
One-side GT due to a combination of HR and NHEJ. Using GT-mediated knock-in as an example, the Cas9 targeting site is removed by modifying the PAM (a). When DSBs are generated, the initial 3′ single-stranded DNA copies the sequence from the GT-vector, forming a D-loop (b). If reannealing to the genomic target site by a second HR event occurs, this two-side GT event creates the planned genome modification (c), but if the DNA reanneals to the genomic target site via NHEJ, an unpredictable sequence outcome at one side of the DSB takes place (d). If the initial 3′ DNA end of the GT-vector is invading the target locus, this recombinant fragment might later integrate elsewhere, at an ectopic site in the genome by NHEJ (e)
GT in specific cell types, organs or developmental stages
For plant genome engineering, the major purpose is to obtain heritable genome modifications. Therefore, acquiring ubiquitous genome modifications in any cells of a plant are unnecessary and might even be unfavorable for plant growth due to toxicity of DSBs or the malfunction of target genes. Moreover, ubiquitously-expressed promoters might not be strong promoters in germline cells; for example, the most frequently used 35S promoter in plants is actually lacking strong activity in meristem or reproductive organs in Arabidopsis (Ge et al. 2008). Several applications of using germline-specific promoters for plant genome modifications were reported in recent years. These promoters that induce transcription in the meristem, sporophyte, female gametophyte or male gametophyte were used to drive the expression of site-specific nucleases. Applying egg-cell-specific promoters to induce Cas9-mediated DSBs successfully enhanced the efficiency of mutagenesis (Mao et al. 2016; Wang et al. 2015; Yan et al. 2015). Very recently, two independent studies applied the EC1 transcriptional cassette for performing GT in Arabidopsis and successfully enhanced the efficiency drastically, in comparison with ubiquitous promoters or other germline-specific promoters (Miki et al. 2018; Wolter et al. 2018). The underlying reasons as to why an egg-cell promoter-driven Cas9 exhibits higher efficiencies for both gene editing and GT are not clear; however, it could simply be due to the fact that any stably induced genetic change within the egg-cell genome will be transferred to the germline, with 100% efficiency.
Furthermore, meristem-specific promoters from YAO55, CDC45 and CLAVATA3 (CLV3, AT2G27250) were also compared to the egg-cell promoter, in terms of their performance in GT efficiency (Miki et al. 2018; Wolter et al. 2018). The CLV3 and CDC45 promoters did not produce any inheritable GT events, and although YAO produced few GT events resulting in herbicide resistant seedlings (0.08%), this efficiency was even lower than for the Ubiquitin promoter (0.3%) (Wolter et al. 2018). Taking the results with the egg-cell promoter into account, it is now of particular interest to also test the male gametophyte for GT. Unfortunately, the pollen-specific promoter, Lat52, did not result in any GT events (Miki et al. 2018), and a Lat52 promoter-driven Cas9 did not perform efficient gene editing mutagenesis (Mao et al. 2016). Intriguingly, the expression profiles are somehow parallel between EC1 in female gametophytes and Lat52 in male gametophytes at the mature stage. The GUS reporter lines for the Lat52 promoter showed pollen-specific activity but no activity in immature anthers, which contain only tetrads and mononucleate microspores (Twell et al. 1990). In parallel, EC1-encoded peptide function in sperm cell activation for fertilization in egg cells (Sprunck et al. 2012). These could be taken as a hint that mature female gametophytes may have a more efficient HR DNA repair system than male gametophytes.
Interestingly, detection of homozygous GT seedlings was reported for the use of the egg-cell promoter in the T2 generation (Miki et al. 2018). One possibility for this is that GT happened directly during egg-cell transformation by agrobacteria, in the T0 generation. Compiled evidence suggests egg cells are the primary target of floral-dip mediated transformation in Arabidopsis (Bent 2000). The second possibility is that the EC1 promoter also possesses embryonic activity (Steffen et al. 2007) that might activate GT at the embryonic stage of either T1 or T2 generations, generating biallelic GT plants. However, homozygous GT seedlings have to be examined with care as the detection of homozygous GT events by PCR cannot differentiate between real biallelic GT events and one GT allele plus a large deletion caused by DSB repair. One recent publication pointed out this kind of pitfall in that single allelic GT plus large deletions could be a frequent phenomenon in mouse zygotes (Adikusuma et al. 2018). It is apparent that large deletions occur regularly during DSB repair in Arabidopsis (Kirik et al. 2000), and therefore, a detailed analysis confirming homozygous GT by segregation rates, or ultimately involving quantitative PCR, is deemed necessary.
Activation of the GT-vector by DNA replication and repair
Increasing the copy number of the template DNA is a simple concept to enhance GT; however, in practice this is not so easy. One innovative approach became popular amongst plant scientists during recent years with the application of geminivirus replicons as GT-vectors for HR, initially developed in the group of Dan Voytas (Baltes et al. 2014; Cermak et al. 2015; Dahan-Meir et al. 2018; Gil-Humanes et al. 2017; Hahn et al. 2018; Pater et al. 2018; Wang et al. 2017). The delivery of a modified virus replicon can be achieved by putting the essential proteins and DNA sequences onto a T-DNA and subsequently delivering them via Agrobacteria. In doing this, Rep/RepA proteins, and LIR and SIR sequences can be bought into plant cells. This method was used to target a transcription factor or introducing herbicide resistance in tobacco and tomato (Baltes et al. 2014; Cermak et al. 2015).
In dicots, most studies use bean yellow dwarf virus (BeYDV) as a tool for GT. An early application of geminivirus replicons in tomato used both TALENs and CRISPR/Cas9 to replace the promoter of the transcription factor ANT1 with the strong 35S promoter, resulting in the accumulation of anthocyanin and thus purple callus and seedlings (Cermak et al. 2015). In another study, an extremely high GT efficiency was reported, without the use of a selection marker, causing the restoration of tomato fruit color from mutant to wild type (Dahan-Meir et al. 2018). BeYDV-expressing cassette was also used to restore the function of pre-integrated GUS and NPTII genes in tobacco (Cermak et al. 2017). In monocots, wheat dwarf virus (WDV) was also used for geminivirus-facilitated GT approaches. Using wheat protoplast systems, the GT events can be quantified to around 1% efficiency, without pre-integrated DNA (Gil-Humanes et al. 2017); however, these GT wheat protoplasts were not regenerated to establish intact plants. The application of WDV in rice thus demonstrated the possibility of achieving GT events via geminivirus replicons, using biolistic transformation, although no heritable change was reported (Wang et al. 2017). It seems that there are multiple reasons as to why the enhancement of GT using the viral GT-vector occurs. This can be because in addition to virus replication increasing vector copy numbers, Rep/RepA also influences the cell to enter nuclear DNA replication (Orozco et al. 2000), which may also enhance GT itself. Moreover, replication itself might also activate the DNA template for repair. Recent results indicate that during DNA repair, damaged DNA might be transferred to repair loci at the nuclear periphery, by the action of nuclear actin and myosin (Caridi et al. 2018). This might also be the reason why the “in planta gene targeting” strategy is also improving GT efficiencies, as in this case DSBs are not only induced at the GT locus but are also used for cleaving and releasing the GT-vector from the genome (Fauser et al. 2012; Hahn et al. 2018; Schiml et al. 2014; Wolter et al. 2018). With this being said, two reports indicated that the geminiviral approach might not be applicable to all dicots. For example, in Arabidopsis, all reported approaches failed to significantly enhance GT (Hahn et al. 2018; Pater et al. 2018), despite replication of the BeYDV replicon being detectable.
New technologies applicable to GT in plant
The fast-growing use of the CRISPR/Cas system opens up a new era of applications in genome engineering in various model organisms, from prokaryotes to eukaryotes. The applications of CRISPR/Cas systems in various organisms provide several interesting approaches that are likely going to be also applicable in plants (Puchta 2017).
A recent innovative approach was undertaken in mammalian cells whereby the local concentration of a factor involved in the resection of DSB ends, an initial step of HR, was carried out. If the protein CtIP, or a fragment of it containing its multimerization domain, is fused to the Cas9 nuclease that is inducing the DSB within the target locus, GT frequencies can be doubled (Charpentier et al. 2018). Another novel approach is bringing the GT-vector DNA within close proximity of the target site to increase GT efficiency. Researchers used pre-assembled CRISPR/Cas9, whereby CRISPR/Cas9 is covalently or non-covalently linked to a GT-vector DNA, forming an RNA–protein–DNA complex, to perform GT in mammalian models (Aird et al. 2018; Gu et al. 2018; Ma et al. 2017; Savic et al. 2018). The underlying mechanism can be considered relatively simple. Upon the formation of a DNA DSB via CRISPR/Cas, the searching of a homologous sequence from the break site to the GT-vector, for repair, can be performed more efficiently if the GT-vector DNA is in close proximity to the DSB. One such approach in yeast used the artificial fusion protein, LexA-Fkh1p, to recruit the GT-vector to the DSB, resulting in a fivefold increase in efficiency of GT (Roy et al. 2018), via the action of both the protein–DNA interaction between the LexA protein and the Lex Operator within the GT-vector, and also the capability of Fkh1p recruitment to the DSB. Additionally performed in yeast and demonstrating an improvement in GT was the use of a VirD2–I-SceI fusion protein expressed in Agrobacterium, that can covalently bind to DNA, transferring DNA from bacteria to yeast, generating a DSB via homing endonuclease I-SceI activity (Rolloos et al. 2015). All of these results indicate that bringing the GT-vector within close proximity of the GT site might also have a positive effect on GT efficiency in plants.
In addition to the well-developed CRISPR/Cas9, other CRISPR/Cas systems, such as CRISPR/Cas12a, provide additional advantages for GT techniques. CRISPR/Cas12a, also known as CRISPR/Cpf1, has two different features that seem to be beneficial for GT, compared to Cas9. First, the induced DSB site is close to one end of the protospacer, far away from the PAM sequence. This feature possibly lets the target be cleaved multiple times, thus increasing the chances of GT, without being blocked by mutations induced by NHEJ. Second, Cas12a produces 5′ protruding ends following DSB induction. Indications that 5′ protruding DSB ends, produced by a paired-nickase version of SpCas9, enhance the efficiency of GT in plants and human cell lines to a certain extent, have recently been reported (Bothmer et al. 2017; Cermak et al. 2017; Wolter et al. 2018). Utilizing CRISPR/Cas12a in rice also achieved high GT efficiency of up to 8% (Begemann et al. 2017), although the numbers of GT events obtained were too small to draw general conclusions. This approach is especially efficient when combining the pre-assembled CRISPR/Cas12a and ssDNA as template for GT. In green algae Chlamydomonas reinhardtii, the utilization of Cas12- and ssDNA-composed GT-vector improved the efficiency of precise GT (Ferenczi et al. 2017). Applying Cas12 with ssDNA as GT-vector was successfully carried out in rice as well (Li et al. 2018b); although we need further data to come to a final conclusion as to whether the application of Cas12 offers higher GT efficiency than Cas9.
Conclusion and perspective
In this report, we attempted to summarize the most recent developments that have already substantially improved GT efficiencies in plants. It turns out that the efficiency and nature of the nuclease used, at which organ the promoter is active, and also the nature and activation of the template, are all critical points. All of these improvements have led to GT frequencies above percent ranges, thus making it a feasible technology for application in plants. Combining different approaches, manipulating the repair machinery and bringing the target and vector in close proximity to one another might help to improve efficiencies even further. We are, therefore, optimistic that within the next few years, GT will become a routine technique for use in crop plants, just as it already is at present for various model animal species.
Author contribution statement
All authors wrote, read and approved the final manuscript.
References
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284. https://doi.org/10.1038/nature24049
Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ (2018) Large deletions induced by Cas9 cleavage. Nature 560:E8–E9. https://doi.org/10.1038/s41586-018-0380-z
Aird EJ, Lovendahl KN, St. Martin A, Harris RS, Gordon WR (2018) Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol 1:816. https://doi.org/10.1038/s42003-018-0054-2
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163. https://doi.org/10.1105/tpc.113.119792
Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M (2017) Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep 7:11606. https://doi.org/10.1038/s41598-017-11760-6
Bent AF (2000) Arabidopsis in planta transformation. uses, mechanisms, and prospects for transformation of other species. Plant Physiol 124:1540–1547. https://doi.org/10.1104/pp.124.4.1540
Bothmer A, Phadke T, Barrera LA, Margulies CM, Lee CS, Buquicchio F, Moss S, Abdulkerim HS, Selleck W, Jayaram H, Myer VE, Cotta-Ramusino C (2017) Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat Commun 8:13905. https://doi.org/10.1038/ncomms13905
Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I (2018) Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559:54–60. https://doi.org/10.1038/s41586-018-0242-8
Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232. https://doi.org/10.1186/s13059-015-0796-9
Cermak T, Curtin SJ, Gil-Humanes J, Cegan R, Kono TJY, Konecna E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217. https://doi.org/10.1105/tpc.16.00922
Charpentier M, Khedher AHY, Menoret S, Brion A, Lamribet K, Dardillac E, Boix C, Perrouault L, Tesson L, Geny S, Cian A de, Itier JM, Anegon I, Lopez B, Giovannangeli C, Concordet JP (2018) CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun 9:1133. https://doi.org/10.1038/s41467-018-03475-7
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J 95:5–16. https://doi.org/10.1111/tpj.13932
Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K-i, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S (2006) Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 25:5579–5590. https://doi.org/10.1038/sj.emboj.7601434
Endo M, Mikami M, Toki S (2016) Biallelic gene targeting in rice. Plant Physiol 170:667–677. https://doi.org/10.1104/pp.15.01663
Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA (2011) Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J 68:929–937. https://doi.org/10.1111/j.1365-313X.2011.04741.x
Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H (2012) In planta gene targeting. Proc Natl Acad Sci USA 109:7535–7540. https://doi.org/10.1073/pnas.1202191109
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359. https://doi.org/10.1111/tpj.12554
Ferenczi A, Pyott DE, Xipnitou A, Molnar A (2017) Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Natl Acad Sci USA 114:13567–13572. https://doi.org/10.1073/pnas.1710597114
Ferreira MG, Cooper JP (2004) Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev 18:2249–2254. https://doi.org/10.1101/gad.315804
Ge X, Wang H, Cao K (2008) Transformation by T-DNA integration causes highly sterile phenotype independent of transgenes in Arabidopsis thaliana. Plant Cell Rep 27:1341–1348. https://doi.org/10.1007/s00299-008-0561-6
Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sanchez-Leon S, Baltes NJ, Starker C, Barro F, Gao C, Voytas DF (2017) High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 89:1251–1262. https://doi.org/10.1111/tpj.13446
Gu B, Posfai E, Rossant J (2018) Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol 36:632–637. https://doi.org/10.1038/nbt.4166
Hahn F, Eisenhut M, Mantegazza O, Weber APM (2018) Homology-directed repair of a defective glabrous gene in Arabidopsis with Cas9-based gene targeting. Front Plant Sci 9:424. https://doi.org/10.3389/fpls.2018.00424
Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J (2001) Gene targeting in Arabidopsis. Plant J 28:671–677. https://doi.org/10.1046/j.1365-313x.2001.01183.x
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832. https://doi.org/10.1038/nbt.2647
Kaya H, Mikami M, Endo A, Endo M, Toki S (2016) Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep 6:26871. https://doi.org/10.1038/srep26871
Kim D, Lim K, Kim S-T, Yoon S-H, Kim K, Ryu S-M, Kim J-S (2017) Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol 35:475–480. https://doi.org/10.1038/nbt.3852
Kirik A, Salomon S, Puchta H (2000) Species-specific double-strand break repair and genome evolution in plants. EMBO J 19:5562–5566. https://doi.org/10.1093/emboj/19.20.5562
Kirik A, Pecinka A, Wendeler E, Reiss B (2006) The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18:2431–2442. https://doi.org/10.1105/tpc.106.045088
La Russa MF, Qi LS (2015) The new state of the art: Cas9 for gene activation and repression. Mol Cell Biol 35:3800–3809. https://doi.org/10.1128/MCB.00512-15
Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10:526–529. https://doi.org/10.1016/j.molp.2016.12.001
Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia L (2018a) Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J Integr Plant Biol 60:536–540. https://doi.org/10.1111/jipb.12650
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018b) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:59. https://doi.org/10.1186/s13059-018-1443-z
Li S, Li J, Zhang J, Du W, Fu J, Sutar S, Zhao Y, Xia L (2018c) Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J Exp Bot 69:4715–4721. https://doi.org/10.1093/jxb/ery245
Ma M, Zhuang F, Hu X, Wang B, Wen X-Z, Ji J-F, Xi JJ (2017) Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-avidin/biotin-donor DNA system. Cell Res 27:578–581. https://doi.org/10.1038/cr.2017.29
Mao Z, Bozzella M, Seluanov A, Gorbunova V (2008) DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7:2902–2906. https://doi.org/10.4161/cc.7.18.6679
Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu J-K (2016) Development of germ-line-specific CRISPR–Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532. https://doi.org/10.1111/pbi.12468
Miki D, Zhang W, Zeng W, Feng Z, Zhu J-K (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9:1967. https://doi.org/10.1038/s41467-018-04416-0
Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M (2018) Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13:195–215. https://doi.org/10.1038/nprot.2017.153
Orel N, Kyryk A, Puchta H (2003) Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J 35:604–612. https://doi.org/10.1046/j.1365-313X.2003.01832.x
Orozco BM, Kong L-J, Batts LA, Elledge S, Hanley-Bowdoin L (2000) The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J Biol Chem 275:6114–6122. https://doi.org/10.1074/jbc.275.9.6114
Paszkowski J, Baur M, Bogucki A, Potrykus I (1988) Gene targeting in plants. EMBO J 7:4021–4026
Pater S de, Klemann B, Hooykaas PJJ (2018) True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci Rep 8:3338. https://doi.org/10.1038/s41598-018-21697-z
Puchta H (1998) Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J 13:331–339. https://doi.org/10.1046/j.1365-313X.1998.00035.x
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14. https://doi.org/10.1093/jxb/eri025
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8. https://doi.org/10.1016/j.pbi.2016.11.011
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57:629–637. https://doi.org/10.1387/ijdb.130194hp
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 93:5055–5060. https://doi.org/10.1073/pnas.93.10.5055
Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF (2013) Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res 23:547–554. https://doi.org/10.1101/gr.145557.112
Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, Buckley SM, Seshacharyulu P, Batra SK, Behlke MA, Zeiner SA, Jacobi AM, Izu Y, Thoreson WB, Urness LD, Mansour SL, Ohtsuka M, Gurumurthy CB (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18:92. https://doi.org/10.1186/s13059-017-1220-4
Reiss B, Klemm M, Kosak H, Schell J (1996) RecA protein stimulates homologous recombination in plants. Proc Natl Acad Sci USA 93:3094–3098. https://doi.org/10.1073/pnas.93.7.3094
Reiss B, Schubert I, Kopchen K, Wendeler E, Schell J, Puchta H (2000) RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc Natl Acad Sci USA 97:3358–3363. https://doi.org/10.1073/pnas.97.7.3358
Rolloos M, Hooykaas PJJ, van der Zaal BJ (2015) Enhanced targeted integration mediated by translocated I-SceI during the Agrobacterium mediated transformation of yeast. Sci Rep 5:8345. https://doi.org/10.1038/srep08345
Roy KR, Smith JD, Vonesch SC, Lin G, Tu CS, Lederer AR, Chu A, Suresh S, Nguyen M, Horecka J, Tripathi A, Burnett WT, Morgan MA, Schulz J, Orsley KM, Wei W, Aiyar RS, Davis RW, Bankaitis VA, Haber JE, Salit ML, St Onge RP, Steinmetz LM (2018) Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol 36:512–520. https://doi.org/10.1038/nbt.4137
Saito S, Maeda R, Adachi N (2017) Dual loss of human POLQ and LIG4 abolishes random integration. Nat Commun 8:16112. https://doi.org/10.1038/ncomms16112
Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17:6086–6095. https://doi.org/10.1093/emboj/17.20.6086
Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li Y, Neri D, Robinson MD, Ciaudo C, Hall J, Jinek M, Schwank G (2018) Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife 7:e33761. https://doi.org/10.7554/eLife.33761
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80:1139–1150. https://doi.org/10.1111/tpj.12704
Schindele P, Wolter F, Puchta H (2018) Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett 592:1954–1967. https://doi.org/10.1002/1873-3468.13073
Shaked H, Melamed-Bessudo C, Levy AA (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA 102:12265–12269. https://doi.org/10.1073/pnas.0502601102
Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338:1093–1097. https://doi.org/10.1126/science.1223944
Steffen JG, Kang I-H, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51:281–292. https://doi.org/10.1111/j.1365-313X.2007.03137.x
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305. https://doi.org/10.1111/tpj.13078
Steinert J, Schiml S, Puchta H (2016) Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep 35:1429–1438. https://doi.org/10.1007/s00299-016-1981-3
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L (2016a) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9:628–631. https://doi.org/10.1016/j.molp.2016.01.001
Sun Y, Li J, Xia L (2016b) Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front Plant Sci 7:1928. https://doi.org/10.3389/fpls.2016.01928
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945. https://doi.org/10.1104/pp.15.00793
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun 7:13274. https://doi.org/10.1038/ncomms13274
Tsakraklides V, Brevnova E, Stephanopoulos G, Shaw AJ (2015) Improved gene targeting through cell cycle synchronization. PLoS One 10:e0133434. https://doi.org/10.1371/journal.pone.0133434
Twell D, Yamaguchi J, McCormick S (1990) Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109:705–713
van Kregten M, Pater S de, Romeijn R, van Schendel R, Hooykaas PJJ, Tijsterman M (2016) T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat Plants 2:16164. https://doi.org/10.1038/nplants.2016.164
Wang Z-P, Xing H-L, Dong L, Zhang H-Y, Han C-Y, Wang X-C, Chen Q-J (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144. https://doi.org/10.1186/s13059-015-0715-0
Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu J-K (2017) Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant 10:1007–1010. https://doi.org/10.1016/j.molp.2017.03.002
Wolter F, Puchta H (2018) The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss army knife for plant biologists. Plant J 94:767–775. https://doi.org/10.1111/tpj.13899
Wolter F, Klemm J, Puchta H (2018) Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J 94:735–746. https://doi.org/10.1111/tpj.13893
Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8:1820–1823. https://doi.org/10.1016/j.molp.2015.10.004
Yang D, Scavuzzo MA, Chmielowiec J, Sharp R, Bajic A, Borowiak M (2016) Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep 6:21264. https://doi.org/10.1038/srep21264
Yourik P, Fuchs RT, Mabuchi M, Curcuru JL, Robb GB (2019) Staphylococcus aureus Cas9 is a multiple-turnover enzyme. RNA 25:35–44. https://doi.org/10.1261/rna.067355.118
Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M (2017) Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun 8:66. https://doi.org/10.1038/s41467-017-00124-3
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu J-L, Wang D, Gao C (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35:438–440. https://doi.org/10.1038/nbt.3811
Acknowledgements
We apologize to all colleagues in this field, as due to space limitations, we were not able to cite all relevant reports on the rapidly growing aspects of genome engineering. We are thankful for the funding support from the Ministry of Science and Technology of Taiwan, ROC (MOST 106-2917-I-564-007-A1) and the Bundesministerium für Forschung und Technologie (100334243 SophGenTom). We also acknowledge Amy Whitbread for English editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Communicated by Laurence Tomlinson.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, TK., Puchta, H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep 38, 443–453 (2019). https://doi.org/10.1007/s00299-019-02379-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02379-0