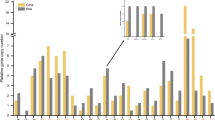
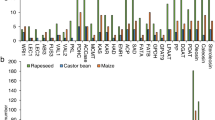
Abstract
Oilseed crops, especially soybean (Glycine max) and canola/rapeseed (Brassica napus), produce seeds that are rich in both proteins and oils and that are major sources of energy and nutrition worldwide. Most of the nutritional content in the seed is accumulated in the embryo during the seed filling stages of seed development. Understanding the metabolic pathways that are active during seed filling and how they are regulated are essential prerequisites to crop improvement. In this review, we summarize various omics studies of soybean and canola/rapeseed during seed filling, with emphasis on oil and protein traits, to gain a systems-level understanding of seed development. Currently, most (80–85%) of the soybean and rapeseed reference genomes have been sequenced (950 and 850 megabases, respectively). Parallel to these efforts, extensive omics datasets from different seed filling stages have become available. Transcriptome and proteome studies have detected preponderance of starch metabolism and glycolysis enzymes to be the possible cause of higher oil in B. napus compared to other crops. Small RNAome studies performed during the seed filling stages have revealed miRNA-mediated regulation of transcription factors, with the suggestion that this interaction could be responsible for transitioning the seeds from embryogenesis to maturation. In addition, progress made in dissecting the regulation of de novo fatty acid synthesis and protein storage pathways is described. Advances in high-throughput omics and comprehensive tissue-specific analyses make this an exciting time to attempt knowledge-driven investigation of complex regulatory pathways.

Similar content being viewed by others
References
Agrawal GK, Thelen JJ (2006) Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics 5:2044–2059
Agrawal GK, Hajduch M, Graham K, Thelen JJ (2008) In-depth investigation of the soybean seed-filling proteome and comparison with a parallel study of rapeseed. Plant Physiol 148:504–518
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Aitken A (2002) Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol 50:993–1010
Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104:16371–16376
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bai F, Settles AM (2015) Imprinting in plants as a mechanism to generate seed phenotypic diversity. Front Plant Sci. doi:10.3389/fpls.2014.00780
Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16:727–741
Borisjuk L, Neuberger T, Schwender J et al (2013) Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell 25:1625–1640
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Brown R, Lemmon B, Nguyen H (2003) Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222:167–174
Chalhoub B, Denoeud F, Liu S et al (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953
Chaudhary J, Patil GB, Sonah H, Deshmukh RK, Vuong TD, Valliyodan B, Nguyen HT (2015) Expanding omics resources for improvement of soybean seed composition traits. Front Plant Sci. doi:10.3389/fpls.2015.01021
Chebrolu KK, Fritschi FB, Ye S, Krishnan HB, Smith JR, Gillman JD (2016) Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 12:1–14
Chen B, Niu F, Liu W-Z et al (2016) Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res 23:101–114
Chrispeels MJ (1991) Sorting of proteins in the secretory system. Annu Rev Plant Biol 42:21–53
Clarke JD, Alexander DC, Ward DP, Ryals JA, Mitchell MW, Wulff JE, Guo L (2013) Assessment of genetically modified soybean in relation to natural variation in the soybean seed metabolome. Sci Rep. doi:10.1038/srep03082
Collakova E, Aghamirzaie D, Fang Y et al (2013) Metabolic and transcriptional reprogramming in developing soybean (Glycine max) embryos. Metabolites 3:347–372
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Demartini DR, Jain R, Agrawal G, Thelen JJ (2011) Proteomic comparison of plastids from developing embryos and leaves of Brassica napus. J Proteome Res 10:2226–2237
Dong J, Keller WA, Yan W, Georges F (2004) Gene expression at early stages of Brassica napus seed development as revealed by transcript profiling of seed-abundant cDNAs. Planta 218:483–491
Du H, Yang S-S, Liang Z, Feng B-R, Liu L, Huang Y-B, Tang Y-X (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. doi:10.1186/1471-2229-12-106
Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16:77–88
Fischer K, Weber A (2002) Transport of carbon in non-green plastids. Trends Plant Sci 7:345–351
Fu X, Fu N, Guo S et al (2009) Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genom. doi:10.1186/1471-2164-10-161
Fulgosi H, Soll J, de Faria Maraschin S, Korthout HA, Wang M, Testerink C (2002) 14-3-3 proteins and plant development. Plant Mol Biol 50:1019–1029
Gajardo HA, Wittkop B, Soto-Cerda B et al (2015) Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol Breeding 35:1–19
Gallardo K, Thompson R, Burstin J (2008) Reserve accumulation in legume seeds. C R Biol 331:755–762
Gan L, C-y Zhang, X-d Wang et al (2013) Proteomic and comparative genomic analysis of two Brassica napus lines differing in oil content. J Proteome Res 12:4965–4978
Garg R, Jain M (2013) RNA-Seq for transcriptome analysis in non-model plants. Methods Mol Biol 1069:43–58
Godovac-Zimmermann J, Kleiner O, Brown LR, Drukier AK (2005) Perspectives in spicing up proteomics with splicing. Proteomics 5:699–709
Goettel W, Liu ZR, Xia J, Zhang WX, Zhao PX, An YQ (2014a) Systems and evolutionary characterization of microRNAs and their underlying regulatory networks in soybean cotyledons. PLoS One. doi:10.1371/journal.pone.0086153
Goettel W, Xia E, Upchurch R, Wang ML, Chen P, An YQ (2014b) Identification and characterization of transcript polymorphisms in soybean lines varying in oil composition and content. BMC Genom. doi:10.1186/1471-2164-15-299
Goldberg RB, De Paiva G, Yadegari R (1994) Plant embryogenesis: zygote to seed. Science 266:605–614
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325:412–421
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38:D843–D846
Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137:1397–1419
Hajduch M, Casteel JE, Hurrelmeyer KE, Song Z, Agrawal GK, Thelen JJ (2006) Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol 141:32–46
Hajduch M, Casteel JE, Tang S, Hearne LB, Knapp S, Thelen JJ (2007) Proteomic analysis of near-isogenic sunflower varieties differing in seed oil traits. J Proteome Res 6:3232–3241
Han J, Lu C, Li Y, Deng Z, Fu B, Geng Z (2016) Discrimination of rapeseeds (Brassica napus L.) based on the content of erucic acid by 1H NMR. Eur Food Res Technol 242:441–447
Harada JJ, Barker SJ, Goldberg RB (1989) Soybean beta-conglycinin genes are clustered in several DNA regions and are regulated by transcriptional and posttranscriptional processes. Plant Cell 1:415–425
Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17:1405–1411
Hardtke CS, Ckurshumova W, Vidaurre DP et al (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131:1089–1100
Harrigan GG, Skogerson K, MacIsaac S, Bickel A, Perez T, Li X (2015) Application of 1H NMR profiling to assess seed metabolomic diversity. A case study on a soybean era population. J Agric Food Chem 63:4690–4697
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Herman EM, Larkins BA (1999) Protein storage bodies and vacuoles. Plant Cell 11:601–613
Hill JF, Breidenbach RW (1974) Proteins of soybean seeds II. Accumulation of the major protein components during seed development and maturation. Plant Physiol 53:747–751
Höglund A-S, Rödin J, Larsson E, Rask L (1992) Distribution of napin and cruciferin in developing rape seed embryos. Plant Physiol 98:509–515
Huang DQ, Koh C, Feurtado JA, Tsang EWT, Cutler AJ (2013) MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genom. doi:10.1186/1471-2164-14-140
Huber SC, Hardin SC (2004) Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol 7:318–322
Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genom. doi:10.1186/1471-2164-15-1
Islam F, Rengifo J, Redden R, Basford K, Beebe SE (2003) Association between seed coat polyphenolics (tannins) and disease resistance in common bean. Plant Foods Hum Nutr 58:285–297
Jin D, Wang Y, Zhao Y, Chen M (2013) MicroRNAs and their cross-talks in plant development. J Genet Genomics 40:161–170
Jofuku KD, Den Boer B, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Jones SI, Vodkin LO (2013) Using RNA-Seq to profile soybean seed development from fertilization to maturity. PLoS One. doi:10.1371/journal.pone.0059270
Joshi T, Yao Q, Franklin LD et al (2010) SoyMetDB: the soybean metabolome database. In: Bioinformatics and biomedicine (BIBM), 2010. IEEE, pp 203–208
Joshi T, Patil K, Fitzpatrick MR et al (2012) Soybean Knowledge Base (SoyKB): a web resource for soybean translational genomics. BMC Genom. doi:10.1186/1471-2164-13-S1-S15
Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98:9448–9453
Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6:4586–4598
Kersten B, Agrawal GK, Iwahashi H, Rakwal R (2006) Plant phosphoproteomics: a long road ahead. Proteomics 6:5517–5528
Kohler C, Lafon-Placette C (2015) Evolution and function of epigenetic processes in the endosperm. Front Plant Sci. doi:10.3389/fpls.2015.00130
Korber N, Bus A, Li J, Parkin IA, Wittkop B, Snowdon RJ, Stich B (2016) Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci. doi:10.3389/fpls.2016.00386
Korbes AP, Machado RD, Guzman F et al (2012) Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS One. doi:10.1371/journal.pone.0050663
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. doi:10.1186/1746-4811-9-29
Kortesniemi M, Vuorinen AL, Sinkkonen J, Yang B, Rajala A, Kallio H (2015) NMR metabolomics of ripened and developing oilseed rape (Brassica napus) and turnip rape (Brassica rapa). Food Chem 172:63–70
Lafon-Placette C, Köhler C (2014) Embryo and endosperm, partners in seed development. Curr Opin Plant Biol 17:64–69
Lam HM, Xu X, Liu X et al (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059
Le BH, Wagmaister JA, Kawashima T, Bui AQ, Harada JJ, Goldberg RB (2007) Using genomics to study legume seed development. Plant Physiol 144:562–574
Lei MG, Reeck GR (1987) Two-dimensional electrophoretic analysis of the proteins of isolated soybean protein bodies and of the glycosylation of soybean proteins. J Agric Food Chem 35:296–300
Li L, Hur M, Lee JY et al (2015) A systems biology approach toward understanding seed composition in soybean. BMC Genom. doi:10.1186/1471-2164-16-S3-S9
Li D, Liu Z, Gao L et al (2016) Genome-wide identification and characterization of microRNAs in developing grains of Zea mays L. PLoS One. doi:10.1371/journal.pone.0153168
Libault M, Farmer A, Joshi T et al (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63:86–99
Lin L, Allemekinders H, Dansby A, Campbell L, Durance-Tod S, Berger A, Jones PJ (2013) Evidence of health benefits of canola oil. Nutr Rev 71:370–385
Lin H, Rao J, Shi J et al (2014) Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J Integr Plant Bio 56:826–836
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Liu X, Huang J, Wang Y, Khanna K, Xie Z, Owen HA, Zhao D (2010) The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J 62:416–428
Liu S, Liu Y, Yang X et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. doi:10.1038/ncomms4930
Lorenz C, Rolletschek H, Sunderhaus S, Braun H-P (2014) Brassica napus seed endosperm—metabolism and signaling in a dead end tissue. J Proteomics 108:382–426
M-a Ohto, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102:3123–3128
M-a Ohto, Floyd SK, Fischer RL, Goldberg RB, Harada JJ (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod 22:277–289
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Meinke D, Chen J, Beachy R (1981) Expression of storage-protein genes during soybean seed development. Planta 153:130–139
Meyer LJ, Gao J, Xu D, Thelen JJ (2012) Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol 159:517–528
Michael TP, VanBuren R (2015) Progress, challenges and the future of crop genomes. Curr Opin Plant Biol 24:71–81
Miller SS, Bowman L-AA, Gijzen M, Miki BL (1999) Early development of the seed coat of soybean (Glycine max). Ann Bot 84:297–304
Moïse JA, Han S, Gudynaitę-Savitch L, Johnson DA, Miki BLA (2005) Seed coats: structure, development, composition, and biotechnology. In Vitro Cell Dev Biol-Pl 41:620–644
Morrell PL, Buckler ES, Ross-Ibarra J (2011) Crop genomics: advances and applications. Nat Rev Genet 13:85–96
Mukherji M (2005) Phosphoproteomics in analyzing signaling pathways. Expert Rev Proteomics 2:117–128
Nakabayashi R, Saito K (2013) Metabolomics for unknown plant metabolites. Anal Bioanal Chem 405:5005–5011
Narvel JM, Fehr WR, Welke GA (1998) Agronomic and seed traits of soybean lines lacking seed lipoxygenases. Crop Sci 38:926–928
Nielsen NC, Dickinson CD, Cho T-J et al (1989) Characterization of the glycinin gene family in soybean. Plant Cell 1:313–328
Niu Y, Wu GZ, Ye R et al (2009) Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana. Mol Plant 2:1107–1122
Nodine MD, Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Gene Dev 24:2678–2692
Norton G, Harris JF (1975) Compositional changes in developing rape seed (Brassica napus L.). Planta 123:163–174
Ohlrogge JB, Kuo T-M (1984) Control of lipid synthesis during soybean seed development: enzymic and immunochemical assay of acyl carrier protein. Plant Physiol 74:622–625
Oikawa A, Nakamura Y, Ogura T et al (2006) Clarification of pathway-specific inhibition by Fourier transform ion cyclotron resonance/mass spectrometry-based metabolic phenotyping studies. Plant Physiol 142:398–413
Oikawa A, Matsuda F, Kusano M, Okazaki Y, Saito K (2008) Rice metabolomics. Rice 1:63–71
Okushima Y, Overvoorde PJ, Arima K et al (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463
O’Rourke JA, Bolon YT, Bucciarelli B, Vance CP (2014) Legume genomics: understanding biology through DNA and RNA sequencing. Ann Bot 113:1107–1120
Pawson T, Scott JD (2005) Protein phosphorylation in signaling—50 years and counting. Trends Biochem Sci 30:286–290
Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13:2777–2791
Peng FY, Weselake RJ (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genom. doi:10.1186/1471-2164-12-286
Petolino JF (2015) Genome editing in plants via designed zinc finger nucleases. In Vitro Cell Dev Pl 51:1–8
Porta H, Rocha-Sosa M (2002) Plant lipoxygenases. Physiological and molecular features. Plant Physiol 130:15–21
Radchuk V, Borisjuk L (2014) Physical, metabolic and developmental functions of the seed coat. Front Plant Sci. doi:10.3389/fpls.2014.00510
Ripp KG, Viitanen PV, Hitz WD, Franceschi VR (1988) Identification of membrane protein associated with sucrose transport into cells of developing soybean cotyledons. Plant Physiol 88:1435–1445
Rubel A, Rinne R, Canvin D (1972) Protein, oil, and fatty acid in developing soybean seeds. Crop Sci 12:739–741
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Schultz DJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis. In: Lipid biotechnology. CRC Press, pp 1–25
Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432:779–782
Severin AJ, Woody JL, Bolon YT et al (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. doi:10.1186/1471-2229-10-160
Shamimuzzaman M, Vodkin L (2012) Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genom. doi:10.1186/1471-2164-13-310
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids 1. Annu Rev Plant Biol 49:611–641
Shewry PR (2009) Wheat. J Exp Bot 60:1537–1553
Smith CW, Patton JG, Nadal-Ginard B (1989) Alternative splicing in the control of gene expression. Annu Rev Genet 23:527–577
Song Q-X, Liu Y-F, Hu X-Y, Zhang W-K, Ma B, Chen S-Y, Zhang J-S (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. doi:10.1186/1471-2229-11-5
Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64:189–217
Swatek KN, Graham K, Agrawal GK, Thelen JJ (2011) The 14-3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J Proteome Res 10:4076–4087
Tan H, Xie Q, Xiang X et al (2015) Dynamic metabolic profiles and tissue-specific source effects on the metabolome of developing seeds of Brassica napus. PLoS One. doi:10.1371/journal.pone.0124794
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4:12–21
Thompson R, Burstin J, Gallardo K (2009) Post-genomics studies of developmental processes in legume seeds. Plant Physiol 151:1023–1029
Thomson DW, Bracken CP, Goodall GJ (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res 39:6845–6853
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Biol 52:335–361
Wang J-W, Wang L-J, Mao Y-B, Cai W-J, Xue H-W, Chen X-Y (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216
Wang X, Wang H, Wang J et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56:253–279
Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10:265–270
Wu G, Zhang L, Yin Y, Wu J, Yu L, Zhou Y, Li M (2015) Sequencing, de novo assembly and comparative analysis of Raphanus sativus transcriptome. Front Plant Sci. doi:10.3389/fpls.2015.00198
Xia Z, Xu H, Zhai J, Li D, Luo H, He C, Huang X (2011) RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol Biol 77:299–308
Xu HM, Kong XD, Chen F, Huang JX, Lou XY, Zhao JY (2015) Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genom. doi:10.1186/s12864-015-2062-7
Ye CY, Xu H, Shen E et al (2014) Genome-wide identification of non-coding RNAs interacted with microRNAs in soybean. Front Plant Sci. doi:10.3389/fpls.2014.00743
Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z (2014) SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genom. doi:10.1186/1471-2164-15-271
Zabala G, Campos E, Varala KK et al (2012) Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biol. doi:10.1186/1471-2229-12-177
Zhang Y, Liang W, Shi J, Xu J, Zhang D (2013) MYB56 encoding a R2R3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J Integr Plant Bio 55:1166–1178
Zhang H, Zhang J, Wei P et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol 12:797–807
Zhang XQ, Sun J, Cao XF, Song XW (2015) Epigenetic mutation of RAV6 affects leaf angle and seed size in rice. Plant Physiol 169:2118–2128
Zhou Z, Jiang Y, Wang Z et al (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:408–414
Acknowledgements
The authors would like to thank Rodrigo Sarria and Steve Rounsley for their support and advice, and Steve Rounsley and Cory Christensen for critical review of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by N. Sreenivasulu.
Pudota B. Bhaskar, Shreedharan Sriram and Po-Hao Wang contributed equally to the review.
Rights and permissions
About this article
Cite this article
Gupta, M., Bhaskar, P.B., Sriram, S. et al. Integration of omics approaches to understand oil/protein content during seed development in oilseed crops. Plant Cell Rep 36, 637–652 (2017). https://doi.org/10.1007/s00299-016-2064-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2064-1