Abstract
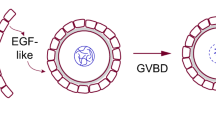
In mammals, female germ cells are sheltered within somatic structures called ovarian follicles, which remain in a quiescent state until they get activated, all along reproductive life. We investigate the sequence of somatic cell events occurring just after follicle activation, starting by the awakening of precursor somatic cells, and their transformation into proliferative cells. We introduce a nonlinear stochastic model accounting for the joint dynamics of the two cell types, and allowing us to investigate the potential impact of a feedback from proliferative cells onto precursor cells. To tackle the key issue of whether cell proliferation is concomitant or posterior to cell awakening, we assess both the time needed for all precursor cells to awake, and the corresponding increase in the total cell number with respect to the initial cell number. Using the probabilistic theory of first passage times, we design a numerical scheme based on a rigorous finite state projection and coupling techniques to compute the mean extinction time and the cell number at extinction time. We find that the feedback term clearly lowers the number of proliferative cells at the extinction time. We calibrate the model parameters using an exact likelihood approach. We carry out a comprehensive comparison between the initial model and a series of submodels, which helps to select the critical cell events taking place during activation, and suggests that awakening is prominent over proliferation.









Similar content being viewed by others
Notes
Although the cut-off r plays a similar role as the index n from Sect. 3.2, we will need two distinct values for the numerical scheme, so that we stick with two different notations, to avoid possible confusion.
We are not able to prove it, as no analytical formula is available for the full model.
We use here the direct simulation rather than Algorithm 1, because the parameter range explored by the symmetric division rate \(\gamma \) gets close to the theoretical necessary and sufficient condition \(\gamma <\alpha _1+\beta \), while Algorithm 1 requires \(2\gamma <\alpha _1+\beta \).
References
Adhikari D, Liu K (2009) Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr Rev 30:438–464
Anderson DF, Kurtz TG (2015) Stochastic analysis of biochemical systems, vol 1. Springer, New York
Bailey N (1964) The elements of stochastic processes. Wiley, New York
Braw-Tal R, Yossefi S (1997) Studies in vivo and in vitro on the initiation of follicle growth in the bovine ovary. J Reprod Fertil 109(1):165–171
Broekmans F, Soules M, Fauser B (2009) Ovarian aging: mechanisms and clinical consequences. Endocr Rev 30(5):465–493
Burnham K, Anderson D (2003) Model selection and multimodel inference: a practical information theoretic approach, 2nd edn. Springer, New York
Cahill LP, Mauleon P (1981) A study of the population of primordial and small follicles in the sheep. J Reprod Fertil 61(1):201–206
Castro M, López-García M, Lythe C, Molina-París C (2018) First passage events in biological systems with non-exponential inter-event times. Sci Rep 8(1):15054
Chou T, D’Orsogna M (2014) First passage problems in biology. In: First-passage phenomena and their applications, pp 306–345. World Scientific
Clément F, Monniaux D (2013) Multiscale modelling of ovarian follicular selection. Prog Biophys Mol Biol 113(3):398–408
Clément F, Michel P, Monniaux D, Stiehl T (2013) Coupled somatic cell kinetics and germ cell growth: multiscale moded-base insight on ovarian follicular development. Multiscale Model Simul 11:719–746
Clément F, Robin F, Yvinec R (2019) Analysis and calibration of a linear model for structured cell populations with unidirectional motion: Application to the morphogenesis of ovarian follicles. SIAM J Appl Math 79(1):207–229
Da Silva-Buttkus P, Jayasooriya G, Mora J, Mobberley M, Ryder T, Baithun M, Stark J, Franks S, Hardy K (2008) Effect of cell shape and packing density on granulosa cell proliferation and formation of multiple layers during early follicle development in the ovary. J Cell Sci 121(23):3890–3900
Darling R, Siegert A (1953) The first passage problem for a continuous markov process. Ann Math Stat 24(4):624–639
Feller W (1967) An introduction to probability theory and its application, vol 1, 3rd edn. Wiley, New York
Fortune J (2003) The early stages of follicular development: activation of primordial follicles and growth of preantral follicles. Anim Reprod Sci 78(3):135–163
Freret-Hodara B, Cui Y, Griveau A, Vigier L, Arai Y, Touboul J, Pierani A (2016) Enhanced abventricular proliferation compensates cell death in the embryonic cerebral cortex. Cereb Cortex
Getto P, Marciniak-Czochra A (2015) Mathematical modelling as a tool to understand cell self-renewal and differentiation. Methods Mol Biol 1293:247–266
Gillespie D (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys 22(4):403–434
Gillespie D (2001) Approximate accelerated stochastic simulation of chemically reacting systems. J Chem Phys 115(4):1716–1733
Glauche I, Cross M, Loeffler R, Roeder I (2007) Lineage specification of hematopoietic stem cells: mathematical modeling and biological implications. Stem Cells 25(7):1791–1799
Gougeon A, Chainy G (1987) Morphometric studies of small follicles in ovaries of women at different ages. J Reprod Fertil 81(2):433–442
Harris T (1963) The theory of branching processes. CRC Press, Berlin
Juengel J, Sawyer H, Smith P, Quirke L, Heath D, Lun S, Wakefield SJ, McNatty K (2002) Origins of follicular cells and ontogeny of steroidogenesis in ovine fetal ovaries. Mol Cell Endocrinol 191(1):1–10
Kimmel M, Axelrod D (2015) Branching processes in biology, vol 19. Springer, New York
Knight P, Glister C (2006) TGF-beta superfamily members and ovarian follicle development. Reproduction 132(2):191–206
Kuntz J (2017) Deterministic approximation schemes with computable errors for the distributions of Markov chains. Ph.D. thesis, Imperial College London
Lintern-Moore S, Moore G (1979) The initiation of follicle and oocyte growth in the mouse ovary. Biol Reprod 20(4):773–778
Lundy T, Smith P, O’connell A, Hudson N, McNatty K (1999) Populations of granulosa cells in small follicles of the sheep ovary. J Reprod Fertil 115(2):251–262
Luria S, Delbrück M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28(6):491–511
Marr C, Strasser M, Schwarzfischer M, Schroeder T, Theis F (2012) Multi-scale modeling of GMP differentiation based on single-cell genealogies. FEBS J 279(18):3488–500
McNatty K, Smith P, Hudson N, Heath D, Tisdall DOW, Braw-Tal R (1995) Development of the sheep ovary during fetal and early neonatal life and the effect of fecundity genes. J Reprod Fertil Suppl 49:123–135
Meredith S, Dudenhoeffer G, Jackson K (2000) Classification of small type B/C follicles as primordial follicles in mature rats. J Reprod Fertil 119(1):43–48
Monniaux D (2016) Driving folliculogenesis by the oocyte-somatic cell dialog: lessons from genetic models. Theriogenology 86(1):41–53
Monniaux D (2018) Factors influencing establishment of the ovarian reserve and their effects on fertility. Anim Reprod 15(Suppl. 1):635–647
Monniaux D, Cadoret V, Clément F, Dalbies-Tran R, Elis S, Fabre S, Maillard V, Monget P, Uzbekova S (2018) Folliculogenesis. In: Huhtaniemi I, Martini L (eds) Encyclopedia of endocrine diseases, 2nd edn. Elsevier, Amsterdam, pp 377–398
Morohaku K (2019) A way for in vitro/ex vivo egg production in mammals. J Reprod Dev 65(4):281–287
Morohaku K, Tanimoto R, Sasaki K, Kawahara-Miki R, Kono T, Hayashi K, Hirao Y, Obata Y (2016) Complete in vitro generation of fertile oocytes from mouse primordial germ cells. Proc Natl Acad Sci USA 113(32):9021–9026
Munsky B, Khammash M (2006) The finite state projection algorithm for the solution of the chemical master equation. J Chem Phys 124(4):044104
Pedersen T (1970) Determination of follicle growth rate in the ovary of the immature mouse. J Reprod Fert 21:81–83
Picton H (2001) Activation of follicle development: the primordial follicle. Theriogenology 55(6):1193–1210
Pujo-Menjouet L (2016) Blood cell dynamics: half of a century of modelling. Math Model Nat Phenom 11(1):92–115
Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, Timmer J (2009) Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25(15):1923–1929
Reader K, Haydon L, Littlejohn R, Juengel J, McNatty K (2012) Booroola BMPR1B mutation alters early follicular development and oocyte ultrastructure in sheep. Reprod Fertil Dev 24(2):353–361
Reddy P, Zheng W, Liu K (2010) Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol Metab 21(2):96–103
Sawyer H, Smith P, Heath D, Juengel J, Wakefield S, McNatty K (2002) Formation of ovarian follicles during fetal development in sheep. Biol Reprod 66(4):1134–1150
Smith POWS, Hudson N, Shaw L, Heath D, Condell L, Phillips D, McNatty K (1993) Effects of the Booroola gene (FecB) on body weight, ovarian development and hormone concentrations during fetal life. J Reprod Fertil 98(1):41–54
Spears N, Molinek M, Robinson L, Fulton N, Cameron H, Shimoda K, Telfer E, Anderson R, Price D (2003) The role of neurotrophin receptors in female germ-cell survival in mouse and human. Development 130(22):5481–5491
Stiehl T, Marciniak-Czochra A (2017) Stem cell self-renewal in regeneration and cancer: insights from mathematical modeling. Methods Mol Biol 5:112–120
Storn R, Price K (1997) Differential evolution—a simple and efficient heuristic for global optimization over continuous spaces. J Global Optim 11:341–359
Tingen C, Kim A, Woodruff T (2009) The primordial pool of follicles and nest breakdown in mammalian ovaries. Mol Hum Reprod 15(2):795–803
Turnbull K, Braden A, Mattner P (1977) The pattern of follicular growth and atresia in the ovine ovary. Aust J Biol Sci 30(3):229–241
Van Kampen N (1992) Stochastic processes in physics and chemistry, vol 1. Elsevier, Amsterdam
Wang C, Zhou B, Xia G (2017) The primordial pool of follicles and nest breakdown in mammalian ovaries. Cell Mol Life Sci 74:2547–2566
Wilkinson D (2011) Stochastic modelling for systems biology. Texts in applied mathematics, 2nd edn. CRC Press, Boca Raton
Wilson T, Wu X, Juengel J, Ross I, Lumsden J, Lord E, Dodds K, Walling G, Mcewan J, O’connell A, Mcnatty K, Montgomery G (2001) Highly prolific booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein Ib receptor (Alk-6) that is expressed in both oocytes and granulosa cells. Biol Reprod 64(4):1225–1235
Zhang H, Risal S, Gorre N, Busayavalasa K, Li X, Shen Y, Bosbach B, Brännström M, Liu K (2014) Somatic cells initiate primordial follicle activation and govern the development of dormant oocytes in mice. Curr Biol 24(21):2501–2508
Acknowledgements
The authors wish to thank Ken McNatty for providing the experimental dataset and Danielle Monniaux for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
1.1 Justification of the choice of the rate of \(\mathcal {R}_2\)
As detailed in Sect. 4.5 the auto-amplification can result from two non-exclusive mechanisms, a nonlocal (global) one and a local one.
Global amplification considers that each proliferative cell sends a fixed amount of growth signals to the oocyte. The oocyte thus receives a signal proportional to the number of proliferative cells C. We consider that the oocyte secrete in turn (instantaneously) a stimulatory signal, at a level proportional to the amount of growth signals received from somatic cells. By homogeneous diffusion, the oocyte signal is shared equally to all somatics cells, so that each precursor cell receives a signal proportional to \(C/(F+C)\).
Local amplification for a given precursor cell, assuming a random repartition of the cell types around the oocyte (hence neglecting local cell-to-cell effects), the probability that a neighbor cell is a proliferative cell is \(C/(F+C-1)\), which is also consistent with our choice.
1.2 Mean-field formulation
To get some insight into the model behavior, we describe the mean-field version of model \(\mathcal {M}_{FC}\), given by the following set of ODE:
with the initial condition \((f(0), c(0)) = (f_0, 0) \), with \(f_0 \in \mathbb {R}_+ \). We start by solving analytically the deterministic formulation, and then investigate the effect of each parameter on the model outputs.
From the ODE system (20), we deduce the change in the proliferative cell proportion \(p_C(t) := \frac{c(t)}{f(t) + c(t)}\):
From ODEs (20) and (21), using the classical method of separation of variables, we can compute the analytical expressions for the proliferative cell proportion \(p_C(t)\), proliferative cell number c(t) and precursor cell number f(t):
Proposition 5
The solution of the ODE system (20) is, for all \(t\ge 0\),
In addition, the solution of ODE (21) is
and the total cell number verifies
From Proposition 5, it is clear that the proliferative cell proportion \(p_C\) converges to 1. If \(\gamma >0\), the proliferative cell number c grows asymptotically exponentially at a rate \(\gamma \) when \(t\rightarrow \infty \). If \(\gamma =0\), c(t) is bounded because \(t\mapsto 1-p_C(t)\) is converging exponentially fast to 0, hence is integrable on \((0,\infty )\). Moreover, the proliferative cell proportion \(p_C\) has an inflexion point if and only if
An inflexion point denotes the presence of at least two distinct phases, with a first progressive acceleration phase followed by a saturating phase.
Finally, note that according to the observed variables, the submodels cannot be distinguished from one another, or, alternatively, different parameter values (within a same submodel) may lead to identical outputs. Indeed, the changes in the precursor cell population are independent of parameters \(\alpha _2,\gamma \), and, more strikingly, parameters \( \beta \) and \(\gamma \) cannot be separated in the analytical solution (22), leading to the same kinetic patterns for \(p_C\) as long as the combination \(\gamma + \beta \) remains unchanged.
1.3 Analytical expressions in the linear case
Proof of Proposition 1
Let \(t \ge 0\) and \(f \in \llbracket 0, f_0 \rrbracket \). Since \(F_t\) is autonomous and is a pure death process, we can directly write the following forward Kolmogorov equation: for all \(f \in \llbracket 0, f_0 \rrbracket \),
Solving by recurrence (23), we deduce that, for all \(f \in \llbracket 0, f_0 \rrbracket \),
Note that \(\mathbb {P}\left[ F_t^L = 0 | F_0 = f_0 \right] = (1 - e^{-\alpha _1 t})^{f_0}\) which converges to 1 when \(t\mapsto 1-p_C(t)\). Hence, process \(F^L\) extincts almost surely (a.s.) when t goes to infinity, hence \(\tau _L< \infty \). Before computing the law of \(\tau _L\), we can directly obtain its mean using the recursive expression (4):
Using again Eq. (4), we deduce that \(\tau _L(= T_{f_0})\) follows a generalized Erlang law whose density function is:
Due to the specific form of the exponential rate, we can simplify Eq. (24) further. As \( \displaystyle \prod _{j \ne i, j = 0}^{f_0 - 1}(f_0 -j) = \frac{f_0 !}{f_0 - i}\) and
we deduce
\(\square \)
Proof of Proposition 2
According to Proposition 1, \(\tau _L\) is a.s. finite. To take the expectation of \(C^L_t \) at time \(t = \tau _L\), we check that \(\mathbb {E}\left[ C^{k,j}_{\tau _L- T_k^j} \right] < \infty \), for all k and j. For all \(t \ge 0\), \(C_t^{k,j}\) is \(L_1-\)integrable (as a Yule process) with \(\mathbb {E}\left[ C^{k,j}_{t} \right] = e^{\gamma t} \). Conditionning on the law of \(\tau _L\), we get (with the change of variables \(x=1-e^{-\alpha _1 t}\))
where B is the standard Beta function. Hence \(I < \infty \) if and only if Hypothesis 3 holds. Note that using the properties of the Beta function, we have
where we use the notation \(\left( m-x\right) !=\prod _{k=1}^m (k-x)\). Thus, if Hypothesis 3 holds true, and given that \(C^{k,j}\) is a positive increasing process, we deduce:
Then, taking the expectation of (6) at time \(t = \tau _L\), we obtain:
Moreover, we have that each counting process \(N_k(t)\) can be dominated by
so that
Finally, conditionally on \(\tau _L\), \(\mathcal {Y}_3(\tau _L)\) is independent of each \(C^{k,j}_{\tau _L}\), and the latter are independent and identically distributed random variables. Using that
and the Wald equation (Feller 1967, Chap. XII), we obtain
which is finite under Hypothesis 3. Finally, if Hypothesis 3 does not hold, we have, as long as \(f_0\ge 2\):
In some special cases, Formula (26) can be used to obtain the first moment of \(C^L_{\tau _L}\).
When \( \gamma \) is zero, then for all \(t \ge 0\), for all \(k \in \llbracket 1, f_0 \rrbracket \) and for all \(j \in \llbracket 1,N_k(\tau _L) \rrbracket \), \( C^{k,j}_t = 1\). We deduce directly from Eq. (26) that
From Eq. (7), we have
by Poisson process property. Since for all \( t \in [T_k, T_{k+1})\), \(F^L_t = f_0 - k\), we deduce that \(\mathbb {E}\left[ N_k(\tau _L) \right] = \mathbb {E}\left[ \alpha _2 (f_0 - k)(T_{k+1} - T_k) \right] \). Using (4), we deduce that \(\mathbb {E} \left[ N_k(\tau _L) \right] = \frac{\alpha _2(f_0 - k) }{\alpha _1(f_0 - k) } = \frac{\alpha _2}{\alpha _1 } \) and conclude with (27).
When \( \alpha _2 \) is zero, \( N_k (t)\) is null for all \(t \ge 0\), and we deduce directly from (26) that
Since \(T_{f_0} = \tau _L\), we have \( C^{f_0,0}_{\tau _L- T_{f_0}} = 1\). Let \(k \in \llbracket 1, f_0 - 1 \rrbracket \). Since \(\tau _L- T_k \overset{(law)}{=} \sum _{i = k + 1}^{f_0} \mathcal {E}\left( \alpha _1 (f_0 - i + 1)\right) \overset{(law)}{=} \sum _{i = 1}^{f_0 - k } \mathcal {E}\left( \alpha _1 i \right) \), using Proposition 1, we deduce that the density function of \(\tau _L- T_k \) is
Then, conditioning \(C^{k,0}_{\tau _L- T_k} \) on the law of \(\tau _L- T_k \), we first deduce that
Then, since \(\mathbb {E} \left[ C^{k,0}_{t} \right] = e^{\gamma t} \), we have, similarly as in Eq. (25),
which ends the proof using (28). \(\square \)
The following proposition is analogous to Proposition 2, yet with the decoupled processes \(\tilde{F}\) and \(\tilde{C}\), whose moments are easier to estimate. Note that parameters \(\tilde{\alpha },\tilde{\beta }, \tilde{\gamma }\) below are generic ones.
Proposition 6
Let \(\tilde{F},\tilde{C}\) be independent pure-jump stochastic processes on \(\mathbb N\), of infinitesimal generators
with deterministic initial condition \(\tilde{F}(0)=f_0\) and \(\tilde{C}(0)=n\ge 1\), and where \(\tilde{\alpha },\tilde{\beta }, \tilde{\gamma }\) are non-negative rate parameters. Let
For any \(p\ge 1\),
if, and only if,
Moreover, we have:
-
if \(\tilde{\gamma } >0\): for \(p=1\),
$$\begin{aligned} \mathbb {E}\left[ \tilde{C}_{\tilde{\tau }} \right] = n\frac{f_0!}{\left( f_0-\frac{\tilde{\gamma }}{\tilde{\alpha }}\right) !}+\frac{\tilde{\beta }}{\tilde{\gamma }} \left( \frac{f_0!}{\left( f_0-\frac{\tilde{\gamma }}{\tilde{\alpha }}\right) !}-1\right) , \end{aligned}$$and for \(p=2\),
$$\begin{aligned} \mathbb {E}\left[ (\tilde{C}_{\tilde{\tau }})^2 \right]= & {} \left( n+\frac{\tilde{\beta }}{\tilde{\gamma }}\right) \left( n+\frac{\tilde{\beta }}{\tilde{\gamma }}+1\right) \frac{f_0!}{\left( f_0-\frac{2\tilde{\gamma }}{\tilde{\alpha }}\right) !}\\&-\left( n+\frac{\tilde{\beta }}{\tilde{\gamma }}\right) \left( 1+2\frac{\tilde{\beta }}{\tilde{\gamma }}\right) \frac{f_0!}{\left( f_0-\frac{\tilde{\gamma }}{\tilde{\alpha }}\right) !}+\left( \frac{\tilde{\beta }}{\tilde{\gamma }}\right) ^2 \end{aligned}$$ -
if \(\tilde{\gamma } = 0\):
$$\begin{aligned} \mathbb {E}\left[ \tilde{C}_{\tilde{\tau }} \right]= & {} n+\frac{\tilde{\beta }}{\tilde{\alpha }} \sum _{i = 1}^{f_0 } \frac{ 1}{i},\\ \mathbb {E}\left[ (\tilde{C}_{\tilde{\tau }})^2 \right]= & {} n+\frac{\tilde{\beta }}{\tilde{\alpha }} \sum _{i = 1}^{f_0 } \frac{ 1}{i}+\frac{\tilde{\beta }^2}{\tilde{\alpha }^2} \left( \sum _{i = 1}^{f_0 } \frac{ 1}{i^2}+\left( \sum _{i = 1}^{f_0 } \frac{ 1}{i}\right) ^2\right) \end{aligned}$$
Proof
Since \(\tilde{\tau } \) and \(\tilde{C}\) are independent, we deduce by conditioning on \(\tilde{\tau } \) that
where \(f_{\tilde{\tau }}\) is the density probability of \(\tilde{\tau } \). Since \(\tilde{F}\) is linear, we apply Proposition 1 and obtain
Now, we suppose that \(\tilde{\gamma } >0 \). Then, \(\tilde{C} \) can be decomposed as the independent sum of n Yule processes starting from 1 [see Eq. (5)] and a birth process with immigration (starting from 0). It is classical that the Yule process follows a geometric law of parameter \(e^{-\tilde{\gamma } t}\), and the birth process with immigration follows a negative binomial law \(\mathcal {BN}\left( \frac{\tilde{\beta }}{\tilde{\gamma }}, e^{- \tilde{\gamma } t} \right) \). Thus there exists \(k,K>0\) (depending on model parameters, but independent of t) such that, for all \(t \ge 0\),
Combining Eq. (33) with Eqs. (31) and (32) yields (29). To obtain the remaining analytical formulas, we note that
and
Also, for any p such that (30) holds true, we have (with the change of variables \(x=1-e^{-\tilde{\alpha } t}\))
where B is the standard Beta function. We deduce that
where we use the notation \(\left( m-x\right) !=\prod _{k=1}^m (k-x)\). Then, using Eqs. (34)-(35) and (32), we deduce from (31) that
and
If \(\tilde{\gamma } = 0\), then \(\tilde{C}\) is a pure immigration process starting from n, and follows a shifted Poisson law \(n+\mathcal {P}\left( \tilde{\beta } t \right) \) at time \(t \ge 0\). Using the same approach, we obtain that
and
. \(\square \)
1.4 Numerical scheme for \(\mathbb {E}[\tau ]\) and \(\mathbb {E}[C_{\tau }] \)
We design Algorithm 1 to compute a numerical estimate of \(g(f_0,0)\), solution of Eq. (13) that represents either \(\mathbb {E}\left[ \tau \right] \) or \( \mathbb {E}\left[ C_{\tau }\right] \) according to the specific choice of boundary condition. This algorithm requires \( \gamma <\alpha _1+\beta \) to compute \(\mathbb {E}\left[ \tau \right] \), and \( 2\gamma <\alpha _1+\beta \) to compute \( \mathbb {E}\left[ C_{\tau }\right] \), in agreement with Theorem 1, Proposition 3 and Proposition 4. The prefactor A given below is obtained thanks to Proposition 6.

1.5 In silico dataset
We generate in silico datasets to further explore parameter identifiability. For each submodel, we choose two different parameter sets with contrasted values in the division rates \(\alpha _2\) or \(\gamma \) and/or transition rate \(\beta \). The parameter values are summarized in Table 2. We obtain the corresponding 10 datasets by simulating 1000 trajectories from the SDE (1), with the Gillespie algorithm (Gillespie 1976), starting from the initial condition \((F_0,0)\) at time \(t=0\) up to the time when \(C(t) = 31\) (the value \(C(t) = 31\) corresponds to the maximal number of cuboidal cells observed in the experimental dataset). The initial random variable \(F_0\) follows a truncated Poisson law of parameter \(\mu \) [see Eq. (17)]. For each trajectory, we select uniformly randomly one point (f, c) among the state space points reached by the trajectory, so that each in silico datasets is composed of \(N=1000\) points. This way of sampling, letting to time-free and uncoupled datapoints, mimics the experimental protocol.
1.6 Detailed fitting procedure
1.6.1 Maximum likelihood estimator
For each submodel and dataset, the optimal parameter values are given by the MLE \(\hat{\theta } = \left( \widehat{\beta } , \widehat{\alpha _2} , \widehat{\gamma }, \widehat{\mu } \right) \), which we compute by minimizing the negative log-likelihood,
for a dataset \(\mathbf {x}\) and where \(\varTheta \) is constructed by fixing all parameters related to the nonpresent events to the singleton \(\{0\}\): for instance, in submodel \((\mathcal {R}_1,\mathcal {R}_4)\), we have \(\varTheta = \{0\} \times \{0\} \times \mathbb {R}_+ \times [1, + \infty )\).
To compute the minimum, we use a derivative-free optimization algorithm: the Differential Evolution (DE) algorithm (Storn and Price 1997). In the following, we describe the whole procedure for the complete model \( (\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3,\mathcal {R}_4)\). The algorithm starts from an initial population in which each individual is represented by a set of real numbers \((\beta , \alpha _2, \gamma , \mu ) \). Then, the population evolves along successive generations by mutation and recombination processes. At each generation, the likelihood function is used to assess the fitness of the individuals, and only the best individuals are kept in the population. We have set the intrinsic optimization parameters as follows: the initial population has a size of 20 individuals, and the probability of mutation and crossing-over equals to 0.8 and 0.7 respectively. The starting individual parameter sets are defined on a log scale, and drawn from a uniform distribution on \(\varTheta = [-6,6]^3 \times [0, 1.5] \). The algorithm was run over 1,000 iterations.
1.6.2 Profile likelihood estimate
For each ith component of the MLE \(\hat{\theta }_i\), \(i \in \llbracket 1, 4 \rrbracket \), we compute a vector \(\hat{\theta }|[\theta _{i} = x]\) on a grid \(G_i\) around the MLE \(\hat{\theta }\), with \(x \in G_i\):
and its associated PLE (vector) \(\mathcal {L}(\mathbf {x};\hat{\theta }|\theta _{i}) \). We design the grid \(G_i\) around the MLE \(\hat{\theta }_{i}\) with a fixed step size (see Table 3 for details), and re-optimize the remaining parameters using the DE algorithm with the same optimization parameters (mut = 0.8, crossp = 0.7, popsize = 20, its = 1000) and initial parameter sets defined on a log scale, and drawn from a uniform distribution on \([-6,6]^3\) for parameters \(\beta \), \(\alpha _2\) and \(\gamma \), and on \([-1 + \log (\hat{\mu }), \log (\hat{\mu }) + 1]\) for parameter \(\mu \).
1.6.3 Confidence intervals
Pointwise likelihood-based confidence intervals are constructed thanks to the likelihood ratio test, following Raue et al. (2009); for each estimated parameter \(\hat{\theta }_{i}\), we select all the parameters \(\theta _{i} = x\) such that:
where \(\varDelta _{0.95} = \chi ^2(0.95, 1) = 3.84\) is the 0.95-quantile of the \(\chi ^2\) law with 1 degree of freedom.
Two-event submodels: Best fit trajectories for in silico datasets. Using Eqs. (15)–(17), we compute each probability \(\mathbb {P}\left[ F_c = f\right] \) for submodel \((\mathcal {R}_1, \mathcal {R}_4)\) (left panel) and \((\mathcal {R}_1, \mathcal {R}_3)\) (right panel) with their respective MLE parameter set associated with the in silico dataset 1. Each dark gray square corresponds to a data point. The colormap corresponds to the probability values \(\mathbb {P}\left[ F_c = f\right] \) in log10 scale (color figure online)
Two-event submodels: PLE for in silico datasets. Each panel represents the PLE, in log10 scale, obtained from the in silico datasets, and either submodel \((\mathcal {R}_1,\mathcal {R}_4)\) (left panels) or \((\mathcal {R}_1,\mathcal {R}_3)\) (right panels). The dashed black line represents the 95%-statistical threshold. Orange solid lines: PLE values for the initial condition parameter \(\mu \); blue solid lines: PLE values for the symmetric cell proliferation rate \(\gamma \); green solid lines: PLE values for the asymmetric cell division rate \(\alpha _2\). The colored points represent the associated MLE, and the star symbols are the expected (true) parameter values (see Table 2) (color figure online)
Three-event submodels: PLE. Each panel represents the PLE, in log10 scale, obtained from the experimental (top panels) and in silico datasets (bottom panels), and either submodel \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_4)\) (left panels), \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3)\) (center panels), \((\mathcal {R}_1, \mathcal {R}_3, \mathcal {R}_4)\) (right panels). The dashed black line represents the 95%-statistical threshold. Orange solid lines: PLE values for the initial condition parameter \(\mu \); blue solid lines: PLE values for the symmetric cell proliferation rate \(\gamma \); green solid lines: PLE values for the asymmetric cell division rate \(\alpha _2\); red solid lines: PLE values for the self-amplification transition rate \(\beta \). The colored points represent the associated MLE, and (in the bottom panels) the star symbols are the expected (true) parameter values (see Table 2) (color figure online)
1.6.4 Model selection
AIC and BIC analyses were performed to compare the submodels. The reader can refer to Burnham and Anderson (2003) (Chapter 6) for a detailed presentation of the rule of thumb, classically used to analyze the \(\varDelta ^{AIC}_i := AIC_i - AIC_{\min }\) and \(\varDelta ^{BIC}_i = BIC_i - BIC_{\min }\) values, where i is the index of the ith model:
-
a \(\varDelta \) value lower than 2 indicates that the considered model is almost as probable as the “best” model;
-
a \(\varDelta \) value between 2 and 7 suggests that the considered model is a suitable alternative to the “best” model;
-
a \(\varDelta \) value between 7 and 10 suggests that the considered model is less relevant than the “best” model;
-
a \(\varDelta \) value upper than 10 suggests that the considered model can be safely ruled out.
This \(\varDelta \) approach is completed by the AIC and BIC weight analyses. For each dataset and criterion (AIC or BIC), we order the AIC/BIC weights from the highest to the lowest values. We then compute the cumulative sum of these weights, starting from the highest one. The selected models are the first ones such that the cumulative sum reaches the threshold p-value 0.95.
1.7 Detailed calibration analysis
1.7.1 Two-event submodels
The fitting results obtained for submodels \((\mathcal {R}_1,\mathcal {R}_3)\) and \((\mathcal {R}_1,\mathcal {R}_4)\) from the experimental datasets are shown in Fig. 5 and discussed in the main text, Sect. 4.3. One fitting result for the in silico datasets and for submodels \((\mathcal {R}_1,\mathcal {R}_3)\) and \((\mathcal {R}_1,\mathcal {R}_4)\) is shown in Fig. 10. We verify that the inferred trajectories are coherent with the selected datasets.
In Fig. 11, we show the PLE for each estimated parameter in each in-silico dataset. Both the initial condition parameter \(\mu \) (orange solid lines) and asymmetric division rate \(\alpha _2 \) (green solid line) are practically identifiable [in the sense given in Raue et al. (2009)], while parameter \(\gamma \) (blue solid line) is only partially practically identifiable in most cases. We observe that both parameters \(\alpha _2\) (\(\mathcal {R}_3\)) and \(\gamma \) (\(\mathcal {R}_4\)) are practically identifiable and close to their expected values (less than one log10 of difference) when the parameters are of the same order of magnitude than \(\alpha _1\). In contrast, a small parameter value compared to \(\alpha _1\) leads to a biased parameter estimate, with a huge shift between the estimated and true parameter values (up to two log10 difference).
The estimator for the initial condition parameter \(\mu \) may also be slightly biased with submodel \((\mathcal {R}_1,\mathcal {R}_3)\) (less than one log10 of difference) compared to submodel \((\mathcal {R}_1,\mathcal {R}_4)\) .
1.7.2 Three-event submodels and complete model
We turn now to the analysis of three-event submodels \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3)\), \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_4)\) and \((\mathcal {R}_1,\mathcal {R}_3,\mathcal {R}_4)\)) and the complete model (\((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3,\mathcal {R}_4)\). Qualitatively, the fitting results for submodel \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3)\) are similar to those for submodel \((\mathcal {R}_1,\mathcal {R}_3)\) (data not-shown); they are characterized by a high probability to produce ten or more proliferative cells before the precursor cell extinction. The fitting results for submodels \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_4)\) and \((\mathcal {R}_1,\mathcal {R}_3,\mathcal {R}_4)\) are rather similar to submodel \((\mathcal {R}_1,\mathcal {R}_4)\); they are characterized by direct cell transition with very little concomitant cell proliferation, followed by prolonged cell proliferation after precursor cell extinction. The fitting results for the complete model are shown in the bottom panels of Fig. 5 for both the Wild-type and Mutant subsets and discussed in the main text, Sect. 4.3.
The PLEs for each dataset and each parameter are presented in Fig. 12 for the three-event submodels. The corresponding parameter values and confidence intervals for the Wild-Type and Mutant subsets are given in Tables 4 and 5. As observed for the two-event submodels, in each case, the initial condition parameter \(\mu \) (orange solid lines) is always practically identifiable, and its fitted value is close to the true one for the in silico datasets. In contrast, all other parameters have a lack of identifiability, both with the experimental and in silico datasets. Specifically, the asymmetric division rate \(\alpha _2\) is practically not identifiable for submodel \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3)\) with the experimental subsets. Interestingly, when the asymmetric division event is combined with the symmetric division event (submodel \((\mathcal {R}_1,\mathcal {R}_3,\mathcal {R}_4)\)) rather than with the auto-amplified transition (submodel \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3)\)), the asymmetric division rate \(\alpha _2\) becomes identifiable in the experimental subsets, which reveals complex parameter dependencies between the asymmetric division rate \(\alpha _2\) and auto-amplified transition rate \(\beta \).
Proliferation versus transition. For the Wild-Type (left panel) and Mutant (right panel) datasets, and for submodel \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_4)\) and complete model \((\mathcal {R}_1,\mathcal {R}_2,\mathcal {R}_3,\mathcal {R}_4)\), we represent in colored lines both the optimal value of self-amplification transition rate \(\beta \) along the PLE of the symmetric cell proliferation rate \(\gamma \), and the optimal value of the symmetric cell proliferation rate \(\gamma \) along the PLE of self-amplification transition rate \(\beta \). The black dashed line represents the straight line \(\gamma =\beta +\alpha _1=\beta +1\) (color figure online)
Rights and permissions
About this article
Cite this article
Clément, F., Robin, F. & Yvinec, R. Stochastic nonlinear model for somatic cell population dynamics during ovarian follicle activation. J. Math. Biol. 82, 12 (2021). https://doi.org/10.1007/s00285-021-01561-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-021-01561-x
Keywords
- Stochastic cell population model
- First passage time
- Finite state projection
- Stochastic coupling techniques
- Maximum likelihood estimate
- Embedded Markov chain