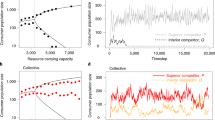
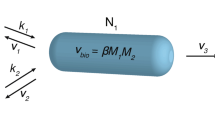
Abstract
Natural selection has shaped the evolution of cells and multi-cellular organisms such that social cooperation can often be preferred over an individualistic approach to metabolic regulation. This paper extends a framework for dynamic metabolic resource allocation based on the maximum entropy principle to spatiotemporal models of metabolism with cooperation. Much like the maximum entropy principle encapsulates ‘bet-hedging’ behaviour displayed by organisms dealing with future uncertainty in a fluctuating environment, its cooperative extension describes how individuals adapt their metabolic resource allocation strategy to further accommodate limited knowledge about the welfare of others within a community. The resulting theory explains why local regulation of metabolic cross-feeding can fulfil a community-wide metabolic objective if individuals take into consideration an ensemble measure of total population performance as the only form of global information. The latter is likely supplied by quorum sensing in microbial systems or signalling molecules such as hormones in multi-cellular eukaryotic organisms.




Similar content being viewed by others
References
Ackermann M (2015) A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13:497–508
An JH, Goo E, Kim H, Seo YS, Hwang I (2014) Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci USA 111:14912–14927
Axelrod R, Hamilton WD (1981) The evolution of cooperation. Science 211:1390–1396
Barrat A, Barthélemy M, Vespignani A (2008) Dynamical processes on complex networks. Cambridge Univ Press, Cambridge
Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T (2015) Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528:99–104
Bélanger M, Allaman I, Magistretti PJ (2011) Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14:724–738
Biggs MB, Papin JA (2013) Novel multiscale modeling tool applied to Pseudomonas aeruginosa biofilm formation. PLoS ONE 8:e78011
Boccaletti S, Bianconi G, Criado R, del Genio CI, Gómez-Gardeñes J, Romance M, deI Sendiña-Nadal I, Wang Z, Zanin M, (2014) The structure and dynamics of multilayer networks. Phys Rep 544:1–122
Boyle KE, Monaco H, van Ditmarsch D, Deforet M, Xavier JB (2015) Integration of metabolic and quorum sensing signals governing the decision to cooperate in a bacterial social trait. PLoS Comput Biol 11:e1004279
Boyle KE, Monaco HT, Deforet M, Yan J, Wang Z, Rhee K, Xavier JB (2017) Metabolism and the evolution of social behavior. Mol Biol Evol 34:2367–2379
Bull JJ, Harcombe WR (2009) Population dynamics constrain the cooperative evolution of cross-feeding. PLoS ONE 4:e4115
Čáp M, Štěpánek L, Harant K, Váchová L, Palková Z (2012) Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 46:436–448
Chao L, Levin BR (1981) Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA 78:6324–6328
Chen J, Gomez JA, Höffner K, Phalak P, Barton PI, Henson MA (2016) Spatiotemporal modeling of microbial metabolism. BMC Syst Biol 10:21
Cole JA, Kohler L, Hedhli J, Luthey-Schulten Z (2015) Spatially-resolved metabolic cooperativity within dense bacterial colonies. BMC Syst Biol 9:15
Davenport PW, Griffin JL, Welch M (2015) Quorum sensing is accompanied by global metabolic changes in the opportunistic human pathogen Pseudomonas aeruginosa. J Bacteriol 197:2072–2082
De Martino D, Andersson AMC, Bergmiller B, Guet CC, Tkac̆ik G, (2017) Statistical mechanics for metabolic networks during steady state growth. Nat Commun 9:2988
D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C (2018) Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep 35:455–488
Estrela S, Gudelj I (2010) Evolution of cooperative cross-feeding could be less challenging than originally thought. PLoS ONE 5:e14121
Evans CR, Kempes CP, Price-Whelan A, Dietrich LEP (2020) Metabolic heterogeneity and cross-feeding in bacterial multicellular systems. Trends Microbiol. https://doi.org/10.1016/j.tim.2020.03.008 (in press)
Fernandez-de-Cossio-Diaz J, Mulet R (2019) Maximum entropy and population heterogeneity in continuous cell cultures. PLoS Comput Biol 15:e1006823
Fernandez-de-Cossio-Diaz J, Mulet R (2020) Statistical mechanics of interacting metabolic networks. Phys Rev E 101:042401
Fleming RMT, Maes CM, Saunders MA, Ye Y, Palsson BØ (2012) A variational principle for computing non-equilibrium fluxes and potentials in genome-scale biochemical networks. J Theor Biol 292:71–77
Françios J, Parrou JL (2001) Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 25:125–145
Gardner A, West SA, Griffin AS (2007) Is bacterial persistence a social trait? PLoS ONE 2:e752
Germerodt S, Bohl K, Lück A, Pande S, Schröter A, Kaleta C, Schuster S, Kost C (2016) Pervasive selection for cooperative cross-feeding in bacterial communities. PLoS Comput Biol 12:e1004986
Goo E, An JH, Kang Y, Hwang I (2015) Control of bacterial metabolism by quorum sensing. Trends Microbiol 23:567–576
Granados AA, Crane MM, Montano-Gutierrez LF, Tanaka RJ, Voliotis M, Swain PS (2017) Distributing tasks via multiple input pathways increases cellular survival in stress. Elife 6:e21415
Grote J, Krysciak D, Streit WR (2015) Phenotypic heterogeneity, a phenomenon that may explain why quorum sensing does not always result in truly homogenous cell behavior. Appl Environ Microbiol 81:5280–5289
Hansen SK, Rainey PB, Haagensen JA, Molin S (2007) Evolution of species interactions in a biofilm community. Nature 445:533–536
Harcombe W (2010) Novel cooperation experimentally evolved between species. Evolution 64:2166–2172
Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, Marx CJ, Segrè D (2014) Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep 7:1104–1115
Harte J, Newman EA (2014) Maximum information entropy: a foundation for ecological theory. Trends Ecol Evol 29:384–389
Hauert C, Doebeli M (2004) Spatial structure often inhibits the evolution of cooperation in the snowdrift game. Nature 428:643–646
Hindmarsh AC, Brown PN, Grant KE, Lee SL, Serban R, Shumaker DE, Woodward CS (2005) SUNDIALS: suite of nonlinear and differential/algebraic equation solvers. ACM Trans Math Softw 31:363–396
Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, Gore J (2016) Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol 14:e1002540
Jaynes ET (1957) Information theory and statistical mechanics. Phys Rev 106:620–630
Jones KD, Kompala DS (1999) Cybernetic model of the growth dynamics of Saccharomyces cerevisiae in batch and continuous cultures. J Biotechnol 71:105–131
Klamt S, Stelling J (2002) Combinatorial complexity of pathway analysis in metabolic networks. Mol Biol Rep 29:233–236
Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078
Lille SH, Pringle JR (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143:1384–1394
Lin YC, Cornell WC, Jo J, Price-Whelan A, Dietrich LEP (2018) The Pseudomonas aeruginosa complement of lactate dehydrogenases enables use of D- and L-lactate and metabolic cross-feeding. mBio 9:e0096-18
Lyssiotis CA, Kimmelman AC (2017) Metabolic interactions in the tumor microenvironment. Trends Cell Biol 27:863–875
Mahadevan R, Edwards JS, Doyle FJ 3rd (2002) Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J 83:1331–1340
Martin W (2010) Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci 365:847–855
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199
Möller P, Liu X, Schuster S, Boley D (2018) Linear programming model can explain respiration of fermentation products. PLoS ONE 13:e0191803
Mori M, Schink S, Erickson DW, Gerland U, Hwa T (2017) Quantifying the benefit of a proteome reserve in fluctuating environments. Nat Commun 8:1225
Morris BE, Henneberger R, Huber H, Moissl-Eichinger C (2013) Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406
Mulcahy LR, Isabella VM, Lewis K (2014) Pseudomonas aeruginosa biofilms in disease. Microb Ecol 68:1–12
Nadell CD, Foster KR, Xavier JB (2010) Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol 6:e1000716
Nadell CD, Drescher K, Foster KR (2016) Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14:589–600
Nakao H, Mikhailov AS (2010) Turing patterns in network-organized activator-inhibitor networks. Nat Phys 6:544–550
Nash J (1950) The bargaining problem. Econometrica 18:155–162
Nelson DL, Cox MM, Michael M (2005) Lehninger principles of biochemistry, 4th edn. W.H, Freeman and Company, New York, p 543
Nowak MA, May RM (1992) Evolutionary games and spatial chaos. Nature 359:826–829
O’Brien EJ, Utrilla J, Palsson BØ (2016) Quantification and classification of E. coli proteome utilization and unused protein costs across environments. PLoS Comput Biol 12:1004998
Oliveira NM, Niehus R, Foster KR (2014) Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci USA 111:17941–17946
Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nat Biotechnol 28:245–248
Othmer HG, Scriven LE (1971) Instability and dynamic pattern in cellular networks. J Theor Biol 32:507–537
Othmer HG, Scriven LE (1974) Nonlinear aspects of dynamic pattern in cellular networks. J Theor Biol 43:83–112
Pacheco AR, Moel M, Segré D (2019) Costless metabolic secretions as drivers of inter-species interactions in microbial ecosystems. Nat Commun 10:103
Pattanaik PK (2008) Social welfare function (definition). In: Durlauf SN, Blume LE (eds) The new Palgrave dictionary of economics, 2nd edn. Palgrave Macmillan, Basingstoke
Perkins TJ, Swain PS (2009) Strategies for cellular decision-making. Mol Syst Biol 5:326
Peterson JR, Cole JA, Luthey-Schulten Z (2017) Parametric studies of metabolic cooperativity in Escherichia coli colonies: strain and geometric confinement effects. PLoS ONE 12:e0182570
Pfeiffer T, Schuster S, Bonhoeffer S (2001) Cooperation and competition in the evolution of ATP-producing pathways. Science 292:504–507
Popat R, Cornforth DM, McNally L, Brown SP (2015) Collective sensing and collective responses in quorum-sensing bacteria. J R Soc Interface 12:20140882
Provost A, Bastin G (2004) Dynamic metabolic modelling under the balanced growth condition. J Process Control 14:717–728
Provost A, Bastin G, Agathos SN, Schneider YJ (2006) Metabolic design of macroscopic bioreaction models: application to Chinese hamster ovary cells. Bioprocess Biosyst Eng 29:349–366
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015:759348
Ridden SJ, Chang HH, Zygalakis KC, MacArthur BD (2015) Entropy, ergodicity, and stem cell multipotency. Phys Rev Lett 115:208103
Schink B (1991) Syntrophism among prokaryotes. In: Ballows AT, Dworkin M, Schleifer KH (eds) The prokaryotes. Springer, New York, pp 276–299
Schink B (2002) Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81:257–261
Schuster S, Hilgetag C (1994) On elementary flux modes in biochemical reaction systems at steady state. J Biol Syst 2:165–182
Seger J, Brockmann HJ (1987) What is bet-hedging? In: Harvey PH, Partridge L (eds) Oxford surveys in evolutionary biology. Oxford University Press, Oxford, pp 182–211
Semenza GL (2008) Tumor metabolism: cancer cells give and take lactate. J Clin Investig 118:3835–3837
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Shore JE, Johnson RW (1980) Axiomatic derivation of the principle of maximum entropy and the principle of minimum cross-entropy. IEEE Trans Inf Theory 26:26–37
Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC (2019) The classification and evolution of bacterial cross-feeding. Front Ecol Evol 7:153
Song HS, Ramkrishna D (2010) Prediction of metabolic function from limited data: Lumped Hybrid Cybernetic Modeling (L-HCM). Biotechnol Bioeng 106:271–284
Song HS, Ramkrishna D (2011) Cybernetic models based on lumped elementary modes accurately predict strain-specific metabolic function. Biotechnol Bioeng 108:127–140
Song H-S, Cannon WR, Beliaev AS, Konopka A (2014) Mathematical modeling of microbial community dynamics: a methodological review. Processes 2:711–752
Sonveaux P et al (2008) Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Investig 118:3930–3942
Tourigny DS (2020) Dynamic metabolic resource allocation based on the maximum entropy principle. J Math Biol 80:2395–2430
Varahan S, Walvekar A, Sinha V, Krishna S, Laxman S (2019) Metabolic constraints drive self-organization of specialized cell groups. Elife 8:e46735
Varma A, Palsson BØ (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Nat Biotechnol 12:994–998
Wolfsberg E, Long CP, Antoniewicz MR (2018) Metabolism in dense microbial colonies: 13C metabolic flux analysis of E. coli T grown on agar identifies two distinct cell populations with acetate cross-feeding. Metab Eng 49:242–247
Woronoff G, Nghe P, Baudry J, Boitard L, Braun E, Griffiths AD, Bibette J (2020) Metabolic cost of rapid adaptation of single yeast cells. Proc Natl Acad Sci USA 117:10660–10666
Young JD, Ramkrishna D (2007) On the matching and proportional laws of cybernetic models. Biotechnol Prog 23:83–99
Young JD, Henne KL, Morgan JA, Konopka AE, Ramkrishna D (2008) Integrating cybernetic modeling with pathway analysis provides a dynamic, systems-level description of metabolic control. Biotechnol Bioeng 100:542–559
Zanghellini J, Gerstl MP, Hanscho M, Nair G, Regensburger G, Müller S, Jungreuthmayer C (2016) Toward Genome-Scale Metabolic Pathway Analysis. In: Wittmann C, Liao JC (eds) Industrial biotechnology
Zhang T, Parker A, Carlson RP, Stewart PS (2018) Flux-balance based modeling of biofilm communities. https://doi.org/10.1101/441311
Acknowledgements
This work benefitted from conversations with HC Causton, L Dietrich, and MR Parsek on microbial biology and quorum sensing. DS Tourigny is a Simons Foundation Fellow of the Life Sciences Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Derivation of the cooperative control law
Appendix A: Derivation of the cooperative control law
Collecting dynamic variables into vectors \({\mathbf {X}}_i = ({\mathbf {m}}_i,x_i)^T\), the system (4) can be represented by the coupled equations \(\dot{{\mathbf {X}}}_i = {\mathbf {F}}_i({\mathbf {X}}, {\mathbf {u}}^i)\) (\(i=1,2,\ldots ,{\mathcal {N}}\)) with \({\mathbf {X}} = ({\mathbf {X}}_1, {\mathbf {X}}_2, \ldots , {\mathbf {X}}_{{\mathcal {N}}})^T\). As in Young and Ramkrishna (2007), Tourigny (2020), the individual controls \({\mathbf {u}}^i\) are determined by using the linearisation of (4) about the state \({\mathbf {X}}(t)\) and reference controls \(\{ {\mathbf {u}}^i_0 \}_{i=1,2,\ldots ,{\mathcal {N}}}\) as a good approximation for the system response at time \(t+\tau \) with \(\tau \in [0,\Delta t]\). The linearised equations are
where
is the ith block row of the full Jacobian matrix \({\mathbf {A}}\) and
with \(\Delta {\mathbf {X}} = {\mathbf {X}}(t+\tau ) - {\mathbf {X}}(t)\) and \(\Delta {\mathbf {u}}^i = {\mathbf {u}}^i (t+ \tau ) - {\mathbf {u}}^i_0\). When the species at all nodes are identical (i.e., \({\mathbf {S}}_i = {\mathbf {S}}\), \(K_i = K\), \({\mathbf {c}}_i = {\mathbf {c}}\), \(r^i_k = r_k\), and \({\mathbf {Z}}^k_i = {\mathbf {Z}}^k\) for all \(i = 1,2,\ldots , {\mathcal {N}}\)) then each cooperative maximum entropy control \({\mathbf {u}}^i\) is obtained by maximising the objective functional
with respect to \({\mathbf {u}}\), where
is the entropy constraint and \(\varvec{\lambda } = (\varvec{\lambda }_1,\varvec{\lambda }_2,\ldots ,\varvec{\lambda }_{{\mathcal {N}}})^T\) is the \({\mathcal {N}} \times (M+1)\)-dimensional vector of Pontryagin co-state variables obtained by solving the boundary value problem
The complete solution to the co-state equations is
but the effective return-on-investment (6) is obtained by substituting for \( \varvec{\lambda }\) with the heuristic \(\tau =0\) because, ultimately, only the optimal control input at the current time t is of interest (Young and Ramkrishna 2007). Maximisation of \({\mathcal {F}}_i\) proceeds as in Tourigny (2020) to yield the cooperative maximum entropy control (7).
An expression for the greedy effective return-on-investment is obtained by setting \(\Delta t = 0\), in which case \(\varvec{\lambda }_i = \partial \phi _i ({\mathbf {X}})/\partial {\mathbf {X}}_i\). Moreover, \(\partial \phi _i/\partial x_i\) is the only non-zero component of this vector because the cooperative objective \(\phi _i\) does not depend on concentrations of slow metabolites. Differentiation of the generalised mean \(\phi _i = M_p({\mathbf {x}})\) with respect to \(x_i\) yields
and the kth column of \({\mathbf {B}}_i\) is
so that
This provides the greedy effective return-on-investment (8) defined using \(R^k_0({\mathbf {m}}_i)\) in the main text, and the resulting greedy cooperative maximum entropy control is
with \(Q_i = \sum _{k=1}^K \exp ( {\mathcal {R}}_0^{k,i} /\sigma )\). As mentioned in the main text, alternative controls can be obtained in cases where the objectives \(\phi _i\) and/or species are different across nodes in the population network, but are not considered here. To demonstrate the utilitarian objective with \(p=1\) recovers the individualistic greedy maximum entropy control law from Tourigny (2020), note that in this case
so that substitution for the denominator in \({\mathcal {R}}^{k,i}_0 \) yields
which is the individualistic effective return-on-investment (Equation 18 in Tourigny 2020) for the kth EFM at node i, multiplied by a common factor of \(1/{\mathcal {N}}\) that has no effect on the resulting control law.
Although beyond the scope of this paper, it is worth pointing out that use of the maximum entropy principle in Tourigny (2020) and its cooperative extension considered here is of a slightly different nature compared to those discussed previously (De Martino et al. 2017; Fernandez-de-Cossio-Diaz and Mulet 2019) (see also Fleming et al. (2012) for explicit use of entropy in an objective function) because the Boltzmann-like distribution is used to weight EFMs rather than individual reactions. The resulting steady state flux distributions obtained using these approaches are not necessarily the same, since the former maximum entropy principle will emphasise the spread of resource across EFMs while the latter does so on a reaction-by-reaction basis. At least in a simplified case where effective return-on-investment is not considered and only stoichiometric constraints must be satisfied, it could be argued the former is more biologically-relevant because spreading resource equally among metabolic pathways– as fundamental functional units of the cell– would be more meaningful than equal distribution of resource among reactions. The latter, albeit done under stoichiometric constraints, could result in metabolic pathways containing more reactions receiving a greater fraction of resource even though their effective return-on-investments are no larger than others.
Rights and permissions
About this article
Cite this article
Tourigny, D.S. Cooperative metabolic resource allocation in spatially-structured systems. J. Math. Biol. 82, 5 (2021). https://doi.org/10.1007/s00285-021-01558-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-021-01558-6
Keywords
- Metabolism
- Elementary flux mode
- Flux balance analysis
- Optimal control
- Maximum entropy
- Information theory
- Social welfare