Abstract
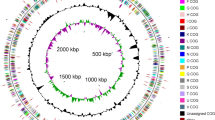
Acinetobacter tandoii SC36 was isolated from a mangrove wetland ecosystem in the Dongzhaigang Nature Reserve in Haikou, China. This bacterium was found to have a capacity for polyphosphate accumulation. To provide insight into its phosphorus metabolism and facilitate its application in phosphorus removal, we developed a draft genome of this strain. KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation revealed three ppk genes and several phosphate metabolic related pathways in the genome of SC36. These genome data of Acinetobacter tandoii SC36 will facilitate elucidation of the mechanism of polyphosphate accumulation.



Similar content being viewed by others
References
Seviour RJ, Mino T, Onuki M (2003) The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol Rev 27(1):99–127
Nakamura K, Masuda K, Mikami E (1991) Isolation of a new type of polyphosphate accumulating bacterium and its phosphate removal characteristics. J Ferment Bioeng 71(4):258–263. https://doi.org/10.1016/0922-338X(91)90278-O
Doughari HJ, Ndakidemi PA, Human IS, Benade S (2011) The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ 26(2):101–112
Tinsley CR, Manjula BN, Gotschlich EC (1993) Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect Immun 61(9):3703–3710
Morohoshi T, Yamashita T, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kuroda A (2003) A method for screening polyphosphate-accumulating mutants which remove phosphate efficiently from synthetic wastewater. J Biosci Bioeng 95(6):637–640
Bond PL, Erhart R, Wagner M, Keller J, Blackall LL (1999) Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol 65(9):4077–4084
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20(2):265–272. https://doi.org/10.1101/gr.097261.109
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23(6):673–679. https://doi.org/10.1093/bioinformatics/btm009
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. https://doi.org/10.1186/1471-2105-4-41
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421. https://doi.org/10.1186/1471-2105-10-421
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428(4):726–731. https://doi.org/10.1016/j.jmb.2015.11.006
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100–1103. https://doi.org/10.1099/ijsem.0.000760
Minkin I, Patel A, Kolmogorov M, Vyahhi N, Pham S (2013) Sibelia: a scalable and comprehensive synteny block generation tool for closely related microbial genomes. In: International workshop on algorithms in bioinformatics. Springer, Berlin, pp 215–229
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31(22):3691–3693. https://doi.org/10.1093/bioinformatics/btv421
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A (2005) Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem 280(2):1448–1456. https://doi.org/10.1074/jbc.M410104200
Bisicchia P, Lioliou E, Noone D, Salzberg LI, Botella E, Hubner S, Devine KM (2010) Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate-limited Bacillus subtilis cells. Mol Microbiol 75(4):972–989. https://doi.org/10.1111/j.1365-2958.2009.07036.x
Creager-Allen RL, Silversmith RE, Bourret RB (2013) A link between dimerization and autophosphorylation of the response regulator PhoB. J Biol Chem 288(30):21755–21769. https://doi.org/10.1074/jbc.M113.471763
Stock JB, Ninfa AJ, Stock AM (1989) Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev 53(4):450–490
Bhutkar A, Russo S, Smith TF, Gelbart WM (2006) Techniques for multi-genome synteny analysis to overcome assembly limitations. Genome Inform 17(2):152–161
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645. https://doi.org/10.1101/gr.092759.109
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64(Pt 2):346–351. https://doi.org/10.1099/ijs.0.059774-0
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Curr Opin Genet Dev 15(6):589–594. https://doi.org/10.1016/j.gde.2005.09.006
Acknowledgements
This study was financially supported by grants from the Natural Science Foundation of China (41366001) and the Education Bureau of Hainan Province (Hnky2017ZD-13). We thank Hainan Dongzhaigang Nature Reserve Authority for assistance with soil and water sample collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that there was no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., Gong, J., Wu, S. et al. Draft genome Sequence of Phosphate-Accumulating Bacterium Acinetobacter tandoii SC36 from a Mangrove Wetland Ecosystem Provides Insights into Elements of Phosphorus Removal. Curr Microbiol 76, 207–212 (2019). https://doi.org/10.1007/s00284-018-1611-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1611-0