Abstract
Two strains of Pseudomonas putida, Psp-LUP and Psp-SPAR, capable of growth on the quinolizidine alkaloids, lupanine and sparteine respectively, were studied here. We report the isolation of Psp-SPAR and the complete genome sequencing of both bacteria. Both were confirmed to belong to P. putida, Psp-LUP close to the type isolate of the species (NBRC14164T) and Psp-SPAR close to strains KT2440 and F1. Psp-SPAR did not grow on lupanine but did contain a gene encoding a putative quinolizidine-17-hydroxylase peptide which exhibited high similarity (76%identity) to the lupanine-17-hydroxylase characterised from Psp-LUP.
Similar content being viewed by others
Introduction
Removal of the neurotoxic alkaloid lupanine, present in the beans of many (“bitter”) varieties of cultivated lupin (Lupinus albus), is required before consumption by humans or livestock. Whilst this can be achieved by prolonged soaking in water, more rapid removal can be mediated via microbial fermentation [1, 2]. A strain of Pseudomonas (Psp-LUP [= DH2001]), capable of growth on lupanine as sole source of carbon and nitrogen, was isolated from soil in Poland by Reifer et al. [3], and its use for the removal of lupin alkaloids was suggested. Reifer et al. [3] referred to this organism as “Pseudomonas lupanini” but it was not formally described. Later examination of this isolate by Hopper et al. [4] found it to form a green-yellow fluorescent pigment when cultivated on King’s B agar, and to exhibit other cytological and biochemical similarities to Pseudomonas putida.
Investigation of the mechanism of lupanine degradation by Psp-LUP showed that the initial step in degradation of the fused heterocyclic ring structure containing tertiary nitrogen atoms (Fig. 1) is hydroxylation in the 17- position to give 17-hydroxylupanine [5]. This step is catalysed by an inducible enzyme [6] which has been fully characterized as PQQ (Pyrroloquinoline quinone)-containing haemoprotein [4]. The enzyme is not an oxygenase but acts as a dehydrogenase to give a double bond which is then hydrated to introduce the hydroxyl group. The gene for the enzyme has been sequenced and active enzyme characterised following its heterologous expression [7, 8].
The dominant alkaloid in the South American lupin (L. mutabilis) is sparteine, also a quinolizidine alkaloid, which differs from lupanine in lacking an oxygen on carbon 2 (Fig. 1). Unlike lupanine which must be purified from lupin beans, sparteine is commercially available and is used by chemists as a chiral ligand in organic syntheses [9].
It is likely that similar pathways are used for the degradation of these two compounds and to pursue further the bacterial catabolism of these, a second strain Pseudomonas (Psp-SPAR) was isolated. As the sequence of the gene for lupanine hydroxylase is known (AJ318095; [7]), it was decided to sequence the genomes of both organisms to see if a similar gene was present in the sparteine-degrading bacterium.
Materials and Methods
Isolation of Pseudomonas putida (Psp-SPAR)
The organism was isolated from a garden compost heap in Aberystwyth, Wales (52.4156,-4.0591) in 2010 by selective culture on mineral medium containing 50 mM K+/Na + phosphate buffer (pH 7.0) containing 4 ml/L of salts solution [10], with 2 mM sparteine as the sole carbon and nitrogen source at 30 °C. It was capable of growth aerobically in liquid medium with sparteine (0.05%, w/v) as sole carbon and nitrogen sources with a generation time of just over an hour. Under these conditions there was no growth on the related compound lupanine.
Extracts of cells were prepared as previously described [4]. Sparteine oxidation was measured using a Clark-type oxygen electrode in a stirred vessel at 30 °C. Reaction mixtures contained in 3.0 mM of 50 mM phosphate buffer (pH 7.0), 1 mg phenazine methosulphate, 1 μmol sparteine and 0.2 ml of crude cell extract. Products were extracted with dichloromethane and examined by GCMS on an HP-5 MS column (25 m x 0.2 mm x 0.33 µm film) with helium as the carrier gas and ionization by electron impact.
DNA Preparation and Genome Sequencing
For DNA extraction, cultures were grown on Nutrient Agar (Oxoid Ltd.). A single colony was resuspended in phosphate-buffered saline (pH7.0) and genomic DNA was purified using an equal volume of SPRI beads and resuspended in EB buffer. The beads were washed with extraction buffer containing lysozyme and RNase A, incubated for 25 min at 37 °C. Proteinase K and RNaseA were added and incubated for 5 min at 65 °C.
DNA was quantified using the Quantit dsDNA HS assay in an Eppendorf AF2200 plate reader (triplicate samples). Genomic DNA libraries were prepared using Nextera XT Library Prep Kit (Illumina, San Diego, USA) following the manufacturer’s protocol with the following modifications: 2 ng DNA instead of 1 ng were used as input, and PCR elongation time was increased to 1 min from 30 s. DNA quantification and library preparation were carried out on a Hamilton Microlab STAR automated liquid handling system (Hamilton Robotics, Reno, NV, USA). Pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina (Roche, Pleasanton, CA, USA) on a Roche light cycler 96 qPCR machine (Roche, Pleasanton, CA, USA). Libraries were sequenced on the Illumina HiSeq using a 250 bp paired end protocol.
Reads were adapter trimmed using Trimmomatic 0.30 (http://www.usadellab.org/) with a sliding window quality cut-off of Q15 [11]. De novo assembly was performed on samples using SPAdes version 3.7 [12] (http://cab.spbu.ru/software/spades/), and contigs were annotated using Prokka 1.11 [13] (http://www.vicbioinformatics.com/software.prokka.shtml).
Genome Analyses
The genomic average nucleotide identity (ANI) [14] was calculated via in silico DNA–DNA hybridization using Kostas Lab web server (http://enve-omics.ce.gatech.edu/g-matrix/) [15]. This estimates the average nucleotide identity using both best hits (one-way ANI) and reciprocal best hits (two-way ANI) between two genomic datasets [16].
Results
Genome Assembly
The draft genome of Psp-LUP comprised 6,457,467 bp with average GC content of 62.3% spread over 88 contigs, with an L50 value of 13 (i.e. The longest 13 contigs together constituted half of the total sequence length, with the 13th largest contig having a size of 156,935 bp [= N50]). The draft genome of Psp-SPAR comprised 6,038,563 bp with average GC content of 61.5%spread over 187 contigs, with an L50 of 28 and N50 of 62,376 bp. Details of sequence and strain deposition are given below.
Identity of Strains
Sequence data from the 16S rRNA locus placed the samples clearly within Pseudomonas putida but with only low resolution (data not shown). Therefore, comparison of the genome sequence data for isolates Psp-LUP and Psp-SPAR alongside a range of other genome sequenced P. putida strains was undertaken using whole genome analysis. Yonezuka et al. [17] recently used whole genome comparisons to re-evaluate the intraspecific taxonomy of P. putida and identified nine sub-groups within the species, in addition to the type isolate. Representatives of these nine groups alongside and the type species confirmed the identity of both organisms as Pseudomonas putida.
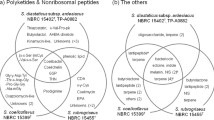
Psp-LUP is closely related (ANI similarity value of 95%; Fig. 2) to the type isolate of P. putida NBRC14164 (= Stanier strain 90 [biotype A] = A.3.12 = ATCC 12,633 = IAM 1236 [18, 19]) which was isolated from soil [20, 21] whilst Psp-SPAR fell clearly within group1 closest to strain F1 (also isolated from soil; [22]). Psp-LUP was closely related to clinical isolates (found in groups 2,3 and 5; Fig. 2) whereas Psp-SPAR clustered with strains isolated from non-clinical sources [23].
Phylogenetic tree (Neighbour-joining) derived from ANI (Average Nuclotide Identity) analysis of 15 P. putida genomes using the grouping designated by Yonezuka et al. (2017). ANI analysis was performed using Kostas Lab web server. Psp-SPAR was clearly placed in group 1, whilst Psp-LUP was closest to NBRC14164T, the type specimen for this species (but which was not assigned to any group by Yonezuka et al.). The tree is rooted using the most distant member of the genus (in group 9). Scalebar indicates percentage sequence divergence
Presence of the 17-Hydroxylase Gene in Ps Putida (SPAR)
A gene was located in the genome of Psp-LUP with a sequence identical to that for lupanine 17-hydroxylase thus confirming the published data for this enzyme [7, 8]. Examination of the genome of Psp-SPAR revealed a very similar gene coding for a protein of 692 amino acid (c.f. 695 for the lupanine 17-hydroxylase in Psp-LUP). Alignment of the two peptides showed them to be very similar (76% identical residues and 86% conserved/identical). Lupanine 17-hydroxylase has a signal sequence of 26 amino acids for transport of the protein into the periplasm. Analysis of the translated gene from Psp-SPAR using SignalP 4.0 [24] also predicted a signal sequence with a cleavage site in this case between residues 27 and 28. Within the protein sequence was the sequence CSGCH at residues 614–618 (c.f. CGACH, 613–617 for lupanine hydroxylase [GenBank Q934G0.1]), corresponding to the haem-binding consensus sequence of CXXCH. Also conserved within the sequence are eight tryptophan-docking motifs that have been shown to be important in the tertiary structure of a number of PQQ-containing proteins [25, 26], including the quinohaemoprotein dehydrogenase from the bacterium Comamonas testosteroni [27], and are also found in lupanine 17-hydroxylase.
Sparteine Degradation by Psp-SPAR
Extracts of cells of Psp-SPAR, grown on sparteine, contained enzymic activity towards sparteine when incubated with phenazine methosulphate as an electron acceptor. This compound, when reduced, is auto-oxidizable and this results in oxygen uptake as measured in an oxygen monitor. Products were identified by GCMS as 17-hydroxysparteine and 17-oxosparteine from their retention times and mass spectra compared with authentic compounds and the NIST library of spectra. 17-hydoxysparteine was prepared by the cold oxidation of sparteine by acid permanganate. 17-Oxosparteine was prepared by the oxidation of sparteine using alkaline ferricyanide as described by Golebiewski and Spenser [28]. No activity was detected in extracts from succinate-grown cells. Psp-LUP was unable to grow on the medium containing sparteine as sole carbon source.
Taxonomic Distribution of Lupanine 17 Hydroxylase Genes
BLASTp searches with putative peptide sequences from Psp-LUP and Psp-SPAR followed by alignment and phylogenetic reconstruction with the most closely related sequences found on GenBank showed these two sequences to form a distinct cluster along with a putative LUH gene (70% identity to the Psp-LUP peptide) from Pseudomonas jesennii, strain GO3 [29], with the next most closely related sequences (from Paraburkholderia sprentiae strain WSM5005, plasmid pl2WSM5005 [438 kb]; [30, 31]) being only 62% identical (Fig. 3). These three putative peptides all contained the haem-binding consensus (CXXCH) and the tryptophan docking motifs. BLASTn searches also did not reveal any more closely related sequences. It is noteworthy that the Paraburkholderia sprentiae containing the putative LUH sequence was isolated from nodules on the roots of the legume Lebeckia ambigua [32]. Like Lupinus spp. Lebeckia spp. contain high levels of both lupanine and sparteine [33].
Phylogenetic tree (Maximum Likelihood tree; Le Gascuel substitution model) of the Psp-LUP and Psp-SPAR peptides alongside the most similar proteins identified by BLASTp searches. Numbers at nodes indicated bootstrap values (as % of 1000 replicates).The tree is rooted using the Sphingomonas histidinilytica quinohemoprotein ethanol dehydrogenase. Scalebar indicates number of substitutions per site
There are over 50 sequenced genomes of P. putida [17] but the LUH gene is not found in any other strains of this species. This and the fact that the LUH genes within Psp-LUH, Psp-SPAR and P. jesennii GO3 are significantly less GC-rich (56.9%, 50.4% and 51.4% respectively) than the genomes in which they occur (all 60-62.3%) suggests that they were acquired by horizontal gene transfer.
Discussion
Thus, as with lupanine metabolism, the first step in sparteine degradation appears to be its hydroxylation on carbon 17 by a dehydrogenase type of enzyme. The 17-oxosparteine product could be produced by a second dehydrogenase reaction catalysed by the same enzyme. This would be analogous to the oxidation of the methyl group of p-cresol first to p-hydroxybenzylalcohol and then to p-hydroxybenzaldehyde by another dehydrogenase-type hydroxylase, the flavocytochrome p-cresol methylhydroxylase [34]. This first step is consistent with the identification of the possible gene for a sparteine 17-hydroxylase from the genome sequencing. These results showing attack on the 17-carbon are also consistent with the finding by Parmaki et al. [1] of 17-oxolupanine as one of the products of bioconversion of lupanine by Pseudomonas putida LPK411.
Accession Numbers
The complete genome sequences were deposited in the European Nucelotide Archive (https://www.ebi.ac.uk/ena) with the accession number for Pseudomonas putida (LUP) of ERX2741807 (PRJEB28091) and for Pseudomonas putida (SPAR) of ERX2741806 (PRJEB28090). Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1).Both cultures are deposited at the National Collection of Industrial, Food and Marine Bacteria (NCIMB Ltd., Aberdeen) with accession numbers 15126 (Psp-LUP) and 15127 (Psp-SPAR) (strains deposited but awaiting accession numbers).
References
Parmaki S, Vyrides I, Vasquez MI, Hartman V, Zacharia I, Hadjiadamou I, Barbeitos CB, Ferreira FC, Afonso CA, Drouza C, Koutinas M (2018) Bioconversion of alkaloids to high-value chemicals: Comparative analysis of newly isolated lupanine degrading strains. Chemosphere 193:50–59
Santana FM, Pinto T, Fialho AM, Sá-Correia I, Empis JM (2002) Bacterial removal of quinolizidine alkaloids and other carbon sources from a Lupinus albus aqueous extract. J Agric Food Chem 50(8):2318–2323
Reifer I, Mozejko-Toczko M (1960) The use of Pseudomonas lupanini in removing alkaloids from bitter lupins. Rocznik Nauk Rolniczych 81(3):711–717
Hopper DJ, Rogozinski J, Toczko M (1991) Lupanine hydroxylase, a quinocytochrome c from an alkaloid-degrading Pseudomonas sp. Biochem J 279(Pt 1):105
Toczko M, Brzeski W, Kakolewska-Baniuk A (1963) Microbial degradation of lupanine. V. Identification of 17-hydroxylupanine. Bull Acad Pol Sci Cl II 11:161–164
Rogoziński J (1975) Molecular properties of the inducible lupanine hydroxylase from growing cultures of Pseudomonas lupanini. Acta Biochim Pol 22(1):57–66
Hopper DJ, Kaderbhai MA, Marriott SA, Young M, Rogozinski J (2002) Cloning, sequencing and heterologous expression of the gene for lupanine hydroxylase, a quinocytochrome c from a Pseudomonas sp. Biochem J 367(2):483–489
Hopper DJ, Kaderbhai MA (2003) The quinohaemoprotein lupanine hydroxylase from Pseudomonas putida. Biochimica et Biophysica Acta (BBA)-Proteins Proteomics 1647(1–2):110–115
Kurek J, Jasiewicz B, Wyrzykiewicz E, Boczoń W (2011) New 2-oxosparteine derivatives: Synthesis and spectroscopic characterization. J Mol Struct 1003(1–3):10–20
Rosenberger R, Elsden S (1960) The yields of Streptococcus faecalis grown in continuous culture. Microbiology 22(3):726–739
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences 106 (45):19126–19131
Rodriguez -RLM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57(1):81–91
Yonezuka K, Shimodaira J, Tabata M, Ohji S, Hosoyama A, Kasai D, Yamazoe A, Fujita N, Ezaki T, Fukuda M (2017) Phylogenetic analysis reveals the taxonomically diverse distribution of the Pseudomonas putida group. The Journal of general applied microbiology 63(1):1–10
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads a taxonomic study. Microbiology 43(2):159–271
Skerman V, McGowan V, Sneath P, Moore W, Moore L (1980) Approved lists. Int J Syst Bacteriol 30:225–420
Den Dooren de Jong L (1926) Bijdrage tot de kennis van het mineralisatieproces. TU Delft, Delft University of Technology
Stanier R (1947) Acetic acid production from ethanol by fluorescent pseudomonads. J Bacteriol 54(2):191
Gibson D, Koch J, Kallio R (1968) Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymic formation of catechol from benzene. Biochemistry 7(7):2653–2662
Molina L, Udaondo Z, Duque E, Fernández M, Bernal P, Roca A, de la Torre J, Ramos JL (2016) Specific gene loci of clinical Pseudomonas putida isolates. PLoS One 11(1):e0147478
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785
Anthony C, Ghosh M (1998) The structure and function of the PQQ-containing quinoprotein dehydrogenases. Progress in biophysics molecular biology 69(1):1–21
Rozeboom HJ, Yu S, Mikkelsen R, Nikolaev I, Mulder HJ, Dijkstra BW (2015) Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenes. A versatile dehydrogenase oxidizing alcohols and carbohydrates. Protein Sci 24(12):2044–2054
Stoorvogel J, Kraayveld DE, Van Sluis CA, Jongejan JA, De Vries S, Duine JA (1996) Characterization of the gene encoding quinohaemoprotein ethanol dehydrogenase of Comamonas testosteroni. Eur J Biochem 235(3):690–698
Golebiewskl WM, Spenser ID (1985) Lactams of sparteine. Can J Chem 63(3):716–719
Otzen M, Palacio C, Janssen DB (2018) Characterization of the caprolactam degradation pathway in Pseudomonas jessenii using mass spectrometry-based proteomics. Appl Microbiol Biotechnol:1–13
Reeve W, Meyer S, Terpolilli J, Melino V, Ardley J, Rui T, Tiwari R, Howieson J, Yates R, O’Hara G (2013) Genome sequence of the Lebeckia ambigua-nodulating “Burkholderia sprentiae” strain WSM5005 T. Standards in genomic sciences 9(2):385
Dobritsa AP, Samadpour M (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66(8):2836–2846
De Meyer SE, Cnockaert M, Ardley JK, Maker G, Yates R, Howieson JG, Vandamme P (2013) Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 63(11):3950–3957
Van Wyk B-E, Verdoorn GH (1989) Chemotaxonomic significance of alkaloids in the genus Lebeckia. Biochem Syst Ecol 17(3):225–229
Hopper DJ, Taylor DG (1977) The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J 167(1):155–162
Acknowledgements
The Institute of Biological, Environmental, and Rural Sciences receives strategic funding from the BBSRC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Detheridge, A.P., Griffith, G.W. & Hopper, D.J. Genome Sequence Analysis of Two Pseudomonas putida Strains to Identify a 17-Hydroxylase Putatively Involved in Sparteine Degradation. Curr Microbiol 75, 1649–1654 (2018). https://doi.org/10.1007/s00284-018-1573-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1573-2