Abstract
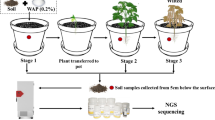
The microbial diversity and the monthly fluctuations in Medicago sativa field soil in response to hydrogen gas were investigated. Illumina high-throughput sequencing was used to analyze the bacteria in raw and hydrogen-treated rhizosphere soil. Among the 18 soil samples, the abundance change of the predominant phyla Proteobacteria and Actinobacteria showed opposite trends. The diversity index analysis of the nine leguminous soil samples showed the highest diversity of the microbial community in July. In the other nine soil samples treated with hydrogen, the microbial diversity decreased and the diversity of soil microorganisms in September was higher than that in July, but not significantly so. The heat map analysis revealed that the microbial community composition of the soil samples was different before and after the hydrogen treatment. After the soil samples were treated with hydrogen, the dominant genera were Nocardioide, Pseudomonas, Janibacter, Microbacterium, Microvirga, Streptomyces, and Phenylobacterium in May; Bradyrhizobium, Haliangium, Sphingomonas, Blastocatella, Lysobacter, and Sphingopyxis in July; and Aeromicrobium, Pseudonocardia, Lentzea, and Skermanella in September. This study indicates that time and hydrogen gas have significant effects on the diversity of microbes in M. sativa rhizospheric soil.



Similar content being viewed by others
References
Berney M, Greening C, Hards K et al (2014) Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol 16:318–330. https://doi.org/10.1111/1462-2920.12320
Bloem J, Veninga M, Sheppard J (1995) Fully automated determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl Environ Microbiol 61:926–936
Bossio DA, Scow KM, Gunapala N et al (1998) Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microbial Ecol 36:1–12. https://doi.org/10.1007/s002489900087
Constant P, Chowdhury SP, Pratscher J et al (2010) Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ Microbiol 12:821–829. https://doi.org/10.1111/j.1462-2920.2009.02130.x
Constant P, Poissant L, Villemur R (2008) Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. Isme J 2:1066. https://doi.org/10.1038/ismej.2008.59
Dong Z, Layzell DB (2001) H2 oxidation, O2 uptake and CO2 fixation in hydrogen treated soils. Plant Soil 229:1–12. https://doi.org/10.1023/A:1004810017490
Dong Z, Wu L, Kettlewell B et al (2003) Hydrogen fertilization of soils—is this a benefit of legumes in rotation? Plant Cell Environ 26:1875–1879. https://doi.org/10.1046/j.1365-3040.2003.01103.x
Duan Q, Liao TW (2013) A new age-based replenishment policy for supply chain inventory optimization of highly perishable products. Int J Pro Econ 145:658–671. https://doi.org/10.1016/j.ijpe.2013.05.020
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Feest A, Madelin MF (1988) Seasonal population changes of myxomycetes and associated organisms in five non-woodland soils, and correlations between their numbers and soil characteristics. Fems Microbiol Lett 53:141–152. https://doi.org/10.1111/j.1574-6968.1988.tb02658.x
George E, Marschner H, Jakobsen I (1995) Role of arbuscular mycorrhizal fungi in uptake of phosphorus and nitrogen from soil. Crit Rev Biotechnol 15:257–270. https://doi.org/10.3109/07388559509147412
Greening C, Berney M, Hards K et al (2014) A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA 111:4257–4261. https://doi.org/10.1073/pnas.1320586111
Grice EA, Kong HH, Conlan S et al (2009) Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. https://doi.org/10.1126/science.1171700
He X, Zou Y, Flynn B et al (2014) Assessment of soil bacterial community structure changes in response to hydrogen gas released by N2-fixing nodules. Int J Plant Soil Sci 3:47–61
Irvine P, Smith M, Dong Z (2004) Hydrogen fertilizer: bacteria or fungi? Acta Hort 631:239–242. https://doi.org/10.17660/ActaHortic.2004.631.30
Kirk JL, Beaudette LA, Hart M et al (2004) Methods for studying soil microbial diversity. J Microbiol Meth 58:169–188. https://doi.org/10.1016/j.mimet.2004.04.006
Lebbink G, Van Faassen HG, Van Ouwerkerk C et al (1994) The Dutch programme on soil ecology of arable farming systems: farm management monitoring programme and general results. Agr Ecosyst Environ 51:7–20. https://doi.org/10.1016/0167-8809(94)90032-9
Lipson DA, Schmidt SK (2004) Seasonal changes in an alpine soil bacterial community in the colorado rocky mountains. Appl Environ Micro 70:2867–2879. https://doi.org/10.1128/AEM.70.5.2867-2879.2004
Lupwayi NZ, Arshad MA, Rice WA, Clayton GW (2001) Bacterial diversity in water-stable aggregates of soils under conventional and zero tillage management. Appl Soil Ecol 16:251–261. https://doi.org/10.1016/S0929-1393(00)00123-2
Mouradi M, Farissi M, Bouizgaren A et al (2016) Effects of water deficit on growth, nodulation and physiological and biochemical processes in Medicago sativa-rhizobia symbiotic association. Arid Land Res Mana 30:193–208. https://doi.org/10.1080/15324982.2015.1073194
Noguchi K, Sakata T, Mizoguchi T et al (2005) Estimating the production and mortality of fine roots in a Japanese cedar (Cryptomeria japonica D. Don) plantation using a minirhizotron technique. J For Res Jpn 10:435–441. https://doi.org/10.1007/s10310-005-0163-x
Orlando J, Chavez M, Bravo L et al (2007) Effect of Colletia hystrix (Clos), a pioneer actinorhizal plant from the Chilean matorral, on the genetic and potential metabolic diversity of the soil bacterial community. Soil Biol Biochem 39:2769–2776. https://doi.org/10.1016/j.soilbio.2007.05.024
Osborne CA, Peoples MB, Janssen PH (2010) Detection of a reproducible, single-member shift in soil bacterial communities exposed to low levels of hydrogen. Appl Environ Microbiol 76:1471–1479. https://doi.org/10.1128/AEM.02072-09
Piche-Choquette S, Tremblay J, Tringe SG, Constant P (2016) H2-saturation of high affinity H2-oxidizing bacteria alters the ecological niche of soil microorganisms unevenly among taxonomic groups. Peer J 4:e1782. https://doi.org/10.7717/peerj.1782
Rumberger A, Merwin IA, Thies JE (2007) Microbial community development in the rhizosphere of apple trees at a replant disease site. Soil Biol Biochem 39:1645–1654. https://doi.org/10.1016/j.soilbio.2007.01.023
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537. https://doi.org/10.1128/AEM.01541-09
Schnurer J, Clarholm M, Rosswall T (1986) Fungi, bacteria and protozoa in soil from four arable cropping systems. Biol Fert Soils 2:119–126. https://doi.org/10.1007/BF00257590
Schutter ME, Dick RP (2002) Microbial community profiles and activities among aggregates of winter fallow and cover-cropped soil. Soil Sci Soc Am J 66:142–153. https://doi.org/10.2136/sssaj2002.1420
Smit E, Leeflang P, Gommans S et al (2001) Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl Environ Microb 67:2284–2291. https://doi.org/10.1128/AEM.67.5.2284-2291.2001
Stark JM (1996) Modeling the temperature response of nitrification. Biogeochemistry 35:433–445. https://doi.org/10.1007/BF02183035
Stursová M, Zifčáková L, Leigh MB et al (2012) Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. Fems Microbiol Ecol 80:735–746
Wang Y, Sheng H, He Y et al (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microb 78:8264–8271. https://doi.org/10.1128/AEM.01821-12
Zhang X, Tian X, Ma L et al (2015) Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. Journal of Biosciences Medicines 03:23–28. https://doi.org/10.4236/jbm.2015.36004
Zou Y (2012) Effect of H2 on soil bacterial community structure and gene expression. Dissertation, Saint Mary’s University, Halifax, NS, Canada
Acknowledgements
This study is supported by the National Natural Science Foundation (Grant No. 41571243) and the project of young talents of science and technology association of Shaanxi Province (Project Number: 20160232). We thank Biomarker Technologies for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, Z., Liu, X., Liu, R. et al. Insight into Bacterial Community Diversity and Monthly Fluctuations of Medicago sativa Rhizosphere Soil in Response to Hydrogen Gas Using Illumina High-Throughput Sequencing. Curr Microbiol 75, 1626–1633 (2018). https://doi.org/10.1007/s00284-018-1569-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1569-y