Abstract
Intestinal microbiota plays a crucial role in preventing the colonization and invasion by pathogens, and disruption of microbiota may cause opportunistic infections and diseases. Pathogens often have strategies to escape from the colonization resistance mediated by microbiota, but whether they also modulate the microbiota composition is still a topic of investigation. In the present study, we addressed this question using an opportunistic pathogen, Klebsiella pneumoniae serotype K1, which is known to cause pyogenic liver abscess (KLA) in about 30% of mice. We examined the effect of K. pneumoniae infection on cecal microbiota composition by performing high-throughput 454 pyrosequencing of the hypervariable V3–V4 regions of bacterial 16S rRNA gene. Our data revealed that K. pneumoniae inoculation substantially changed the cecal microbiota composition when analyzed at the phylum, order, and family levels. Most strikingly, the KLA-infected mice had significantly increased abundance of Bacteroidales and Enterobacteriales and decreased abundance of Lactobacillales and Eggerthellales. Furthermore, by comparing the infected mice with or without KLA disease symptoms, we identified specific microbiota changes associated with the KLA disease induction. Especially, the KLA group had dramatically decreased sequence identical to Lactobacillus compared with non-KLA mice. These findings suggest that the pathogenic process of KLA infection may involve alteration of microbiota compositions, particularly reduction in Lactobacillus.




Similar content being viewed by others
References
Bernet-Camard MF, Liévin V, Brassart D et al (1997) The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl Environ Microbiol 63:2747–2753
Britton RA, Young VB (2014) Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146:1547–1553
Chen N, Wang LL, Xue J et al (2014) Different metabolic profiles of K1 serotype and non-serotype K1 and K2 Klebsiella pneumoniae isolates in oral infection mice model. Microb Pathog 75:41–48
Chung DR, Lee SS, Lee HR et al (2007) Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect 54:578–583
Claesson MJ, Van Sinderen D, O’Toole PW (2008) Lactobacillus phylogenomics-towards a reclassifcation of the genus. Int J Syst Evol Microbiol 58:2945–2954
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Gopal PK, Prasad J, Smart J et al (2001) In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifdobacteriumlactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol 67:207–216
He M, Fang S, Huang X et al (2016) Evaluating the contribution of gut microbiota to the variation of porcine fatness with the cecum and fecal samples. Front Microbiol 7:2108
Hudault S, Lievin V, Bernet-Camard MF et al (1997) Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella typhimurium C5 infection. Appl Environ Microbiol 63:513–518
Kuo SH, Lee YT, Li CR et al (2013) Mortality in emergency department sepsis score as a prognostic indicator in patients with pyogenic liver abscess. Am J Emerg Med 31:916–921
Lin YC, Lu MC, Lin C et al (2013) Activation of IFN-γ/STAT/IRF-1 in hepatic responses to Klebsiella pneumoniae Infection. PLoS ONE 8:e79961
Ling Z, Liu X, Jia X et al (2014) Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep 4:7485
Ling Z, Liu X et al (2015) Decreased diversity of the oral microbiota of patients with hepatitis B virus-induced chronic liver disease: a pilot project. Sci Rep 5:17098
Lynch SV, Pedersen O (2016) The human intestinal microbiome in health and disease. New Engl J Med 375:2369–2379
Roderburg C, Luedde T (2014) The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes 5:441–445
Sabatino A, Regolisti G, Cosola C et al (2017) Intestinal microbiota in Type 2 diabetes and chronic kidney disease. Curr Diab Rep 17:16
Salminen MK, Tynkkynen S, Rautelin H et al (2002) Lactobacillus bacteremia during a rapid increase in probiotic use of Lactobacillus rhamnosus GG in Finland. Clin Infect Dis 35:1155–1160
Sullivan A, Nord CE (2006) Probiotic lactobacilli and bacteraemia in Stockholm. Scand J Infect Dis 38:327–331
Swidsinski A, Loening-Baucke V, Lochs H et al (2005) Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol 11:1131–1140
Tu YC, Lu MC, Chiang MK et al (2009) Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 77:2657–2671
Turton JF, Englender H, Gabriel SN et al (2007) Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol 56:593–597
Vila A, Cassata A, Pagella H et al (2011) Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol J 5:107–113
Zaborin A, Krezalek M, Hyoju S et al (2017) Critical role of microbiota within cecal crypts on the regenerative capacity of the intestinal epithelium following surgical stress. Am J Physiol Gastrointest Liver Physiol 312:G112–G122
Acknowledgements
We thank the support from Affiliated Hospital of Hebei University Postdoctoral workstation and Hebei University Biology Postdoctoral research station. We also thank the support from Key Bioengineering Discipline of Hebei Province.
Funding
This study was sported by Natural Science Foundation of Hebei Province (No. H2015201076), National Natural Science Foundation of China (No. 81000760), Medical Construction Special Project of Hebei University (No. 2014A2003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Nan Chen and Zong-Xin Ling have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2018_1471_MOESM1_ESM.tif
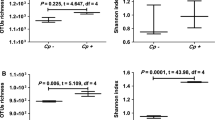
Richness estimators ACE (a) and Chao1 (b) and diversity indices Shannon (c) and Simpson (d) were calculated using mothur. (healthy vs KLA, P > 0.05; non-KLA vs KLA, P > 0.05). (TIF 5701 KB)
284_2018_1471_MOESM2_ESM.tif
Differences in gut microbiota taxon abundance between healthy and non-KLA groups. Comparison of relative abundance in bacterial sequences at the levels of phylum (a), order (b) and family (c) between healthy (Green) and non-KLA (Blue) groups; *P < 0.05. (TIF 7140 KB)
Rights and permissions
About this article
Cite this article
Chen, N., Ling, ZX., Jin, TT. et al. Altered Profiles of Gut Microbiota in Klebsiella pneumoniae-Induced Pyogenic Liver Abscess. Curr Microbiol 75, 952–959 (2018). https://doi.org/10.1007/s00284-018-1471-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1471-7