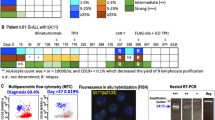
Abstract
Molecular measurable residual disease (MRD) monitoring based on real-time quantitative reverse transcription PCR (RT-qPCR) plays an important role in acute promyelocytic leukemia (APL) management, but the performance status of clinical reports is unknown. This study focuses on the specific elements in molecular MRD monitoring report and their impact on clinical decision-making. The participating laboratories were asked to submit real and formal clinical reports for mock samples panel with APL clinical case. The MRD-specific elements were analyzed and summarized. The significance of longitudinal MRD monitoring curve and the missing MRD-specific elements for clinical decision-making were assessed. MRD-specific elements were significantly missing in clinical reports. The element “testing results” existed great inconsistencies in the written form of testing items and data. The longitudinal MRD monitoring curve of false-negative or false-positive MRD result was obviously different from all-correct. It not only identified MRD time point of tissue sampling relative to treatment and ensured the reliability of the negative MRD results, but also gave MRD diagnosis, clinical interpretation, and further recommendation. Clinician-friendly reports with MRD-specific elements can better serve clinical practice. The correctly intuitive results and clinically important MRD-specific elements can provide a good description of the reliability and clinical significance of MRD results.


Similar content being viewed by others
References
Tallman MS, Kwaan HC (1992) Reassessing the hemostatic disorder associated with acute promyelocytic leukemia. Blood 79(3):543–553
Park JH, Qiao B, Panageas KS, Schymura MJ, Jurcic JG, Rosenblat TL, Altman JK, Douer D, Rowe JM, Tallman MS (2011) Early death rate in acute promyelocytic leukemia remains high despite all-trans retinoic acid. Blood 118:1248–1254. https://doi.org/10.1182/blood-2011-04-346437
Grignani F, Ferrucci PF, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, Pelicci PG (1993) The acute promyelocytic leukemia-specific PML-RARα fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 74(3):423–431
O’Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, Bhatt V, Bixby D, Blum W, Coutre SE, de Lima M, Fathi AT, Fiorella M, Foran JM, Gore SD, Hall AC, Kropf P, Lancet J, Maness LJ, Marcucci G, Martin MG, Moore JO, Olin R, Peker D, Pollyea DA, Pratz K, Ravandi F, Shami PJ, Stone RM, Strickland SA, Wang ES, Wieduwilt M, Gregory K, Ogba N (2017) Acute myeloid leukemia, version 3.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw 15(7):926–957. https://doi.org/10.6004/jnccn.2017.0116
Sanz MA, Grimwade D, Tallman MS, Lowenberg B, Fenaux P, Estey EH, Naoe T, Lengfelder E, Buchner T, Dohner H, Burnett AK, Lo-Coco F (2009) Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 113(9):1875–1891. https://doi.org/10.1182/blood-2008-04-150250
Dayton VJ, McKenna RW, Yohe SL et al (2017) Relapsed acute promyelocytic leukemia lacks “classic” leukemic promyelocyte morphology and can create diagnostic challenges. Am J Clin Pathol 147(1):69–76. https://doi.org/10.1093/ajcp/aqw202
Imagawa J (2018) Switching from all-trans retinoic acid to arsenic trioxide for newly diagnosed acute promyelocytic leukemia. Leuk Lymphoma 4:1–3. https://doi.org/10.1080/10428194.2018.1443333
de Thé H, Chen Z (2010) Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer 10(11):775–783. https://doi.org/10.1038/nrc2943
Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R, Diverio D, Jones K, Aslett H, Batson E, Rennie K, Angell R, Clark RE, Solomon E, Lo-Coco F, Wheatley K, Burnett AK (2009) Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol 27(22):3650–3658. https://doi.org/10.1200/JCO.2008.20.1533
Albano F, Mestice A, Pannunzio A et al (2006) The biological characteristics of CD34+ CD2+ adult acute promyelocytic leukemia and the CD34 CD2 hypergranular (M3) and microgranular (M3v) phenotypes. Haematologica 91(3):311–316
Gabert J, Beillard E, Van der Velden VHJ et al (2003) Standardization and quality control studies of ‘real-time’quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia–a Europe Against Cancer program. Leukemia 17(12):2318–2357. https://doi.org/10.1038/sj.leu.2403135
Østergaard M, Nyvold CG, Jovanovic JV, Andersen MT, Kairisto V, Morgan YG, Tobal K, Pallisgaard N, Özbek U, Pfeifer H, Schnittger S, Grubach L, Larsen JK, Grimwade D, Hokland P (2011) Development of standardized approaches to reporting of minimal residual disease data using a reporting software package designed within the European LeukemiaNet. Leukemia 25(7):1168–1173. https://doi.org/10.1038/leu.2011.69
Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, Grimwade D, Haferlach T, Hills RK, Hourigan CS, Jorgensen JL, Kern W, Lacombe F, Maurillo L, Preudhomme C, van der Reijden BA, Thiede C, Venditti A, Vyas P, Wood BL, Walter RB, Döhner K, Roboz GJ, Ossenkoppele GJ (2018) Minimal/measurable residual disease in AML: consensus document from ELN MRD Working Party. Blood 131(12):1275–1291. https://doi.org/10.1182/blood-2017-09-801498
Beillard E, Pallisgaard N, Van der Velden VHJ et al (2003) Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)–a Europe against cancer program. Leukemia 17(12):2474–2486. https://doi.org/10.1038/sj.leu.2403136
Scheuner MT, Hilborne L, Brown J, Lubin, for the members of the RAND IM (2012) A report template for molecular genetic tests designed to improve communication between the clinician and laboratory. Genet Test Mol Biomarkers 16(7):761–769. https://doi.org/10.1089/gtmb.2011.0328
Deans Z, Ahn JW, Bergbaum A, et al. Best practice guidelines for internal quality control in genetic laboratories. 2015
Gulley ML, Braziel RM, Halling KC et al (2007) Clinical laboratory reports in molecular pathology. Arch Pathol Lab Med 131(6):852–863. https://doi.org/10.1043/1543-2165(2007)131[852:CLRIMP]2.0.CO;2
Haga SB, Mills R, Pollak KI, Rehder C, Buchanan AH, Lipkus IM, Crow JH, Datto M (2014) Developing patient-friendly genetic and genomic test reports: formats to promote patient engagement and understanding. Genome Med 6(7):58. https://doi.org/10.1186/s13073-014-0058-6
Claustres M, Kožich V, Dequeker E et al (2014) Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic). Eur J Hum Genet 22(2):160–170. https://doi.org/10.1038/ejhg.2013.125
Dorschner MO, Amendola LM, Shirts BH, Kiedrowski L, Salama J, Gordon AS, Fullerton SM, Tarczy-Hornoch P, Byers PH, Jarvik GP (2014) Refining the structure and content of clinical genomic reports[C]. Am J Med Genet C: Semin Med Genet 166(1):85–92. https://doi.org/10.1002/ajmg.c.31395
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Löwenberg B, Bloomfield CD (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129(4):424–447. https://doi.org/10.1182/blood-2016-08-733196
Zhan S, Li J, Xu R, Wang L, Zhang K, Zhang R (2009) Armored long RNA controls or standards for branched DNA assay for detection of human immunodeficiency virus type. J Clin Microbiol 47(8):2571–2576. https://doi.org/10.1128/JCM.00232-09
Foroni L, Wilson G, Gerrard G, Mason J, Grimwade D, White HE, de Castro DG, Austin S, Awan A, Burt E, Clench T, Farruggia J, Hancock J, Irvine AE, Kizilors A, Langabeer S, Milner BJ, Nickless G, Schuh A, Sproul A, Wang L, Wickham C, Cross NCP (2011) Guidelines for the measurement of BCR-ABL1 transcripts in chronic myeloid leukaemia. Br J Haematol 153(2):179–190. https://doi.org/10.1111/j.1365-2141.2011.08603.x
Nagai S, Nannya Y, Arai S, Yoshiki Y, Takahashi T, Kurokawa M (2010) Molecular or cytogenetic monitoring and preemptive therapy for central nervous system relapse of acute promyelocytic leukemia. Haematologica 95(1):169–171. https://doi.org/10.3324/haematol.2009.015545
Dunnen JT, Dalgleish R, Maglott DR et al (2016) HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 37(6):564–569. https://doi.org/10.1002/humu.22981
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, Q., Zhang, R., Peng, R. et al. Clinician-friendly reports of molecular measurable residual disease monitoring in acute promyelocytic leukemia. Ann Hematol 98, 2347–2355 (2019). https://doi.org/10.1007/s00277-019-03782-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-019-03782-z