Abstract
We propose a method for examining and measuring the complexity of animal social networks that are characterized using association indices. The method focusses on the diversity of types of dyadic relationship within the social network. Binomial mixture models cluster dyadic relationships into relationship types, and variation in the preponderance and strength of these relationship types can be used to estimate association complexity using Shannon’s information index. We use simulated data to test the method and find that models chosen using integrated complete likelihood give estimates of complexity that closely reflect the true complexity of social systems, but these estimates can be downwardly biased by low-intensity sampling and upwardly biased by extreme overdispersion within components. We also illustrate the use of the method on two real datasets. The method could be extended for use on interaction rate data using Poisson mixture models or on multidimensional relationship data using multivariate mixture models.
Significance statement
Animals from many species interact socially with multiple individuals, and these interactions form a social network. Pairs of individuals have social relationships that differ in their strength and type. This social complexity has long interested behavioural biologists, particularly in the context of social cognition. Measuring social complexity, however, presents challenges. We propose a new method for measuring the complexity of animal social networks. Our approach is based on quantifying variation in the strengths of social connections (measured using association indices) which we use to classify different types of pairwise relationships. We, then, use the number, strength and prevalence of these different types of relationships to measure association complexity. Our approach can be used to compare association complexity between populations and/or species. We provide code that researchers can use with their own datasets.
Similar content being viewed by others
Introduction
Social complexity is a much used concept in behavioural ecology (Kappeler 2019, Topical collection on Social complexity). However, definitions vary widely and, often, are not operationalized. Measures of social complexity have been sought and used for a variety of reasons, perhaps most notably to test the social intelligence hypothesis for the evolution of cognition (Kwak et al. 2018; Kappeler 2019, Topical collection on Social complexity) and the social complexity hypothesis for the evolution of communication (Freeberg et al. 2012).
In studies of non-human societies, the term social complexity has primarily been used in two broad ways. First, social complexity is used to describe the number of different types (roles) of individuals that make up a social group (e.g., Blumenstein and Armitage 1998; Groenewoud et al. 2016). Second, social complexity is used to describe the complexity of social relationships among individuals within a social group or population (e.g., Fischer et al. 2017). Recent work has highlighted the importance of considering these two aspects of social complexity separately. These two types of complexity appear to evolve under different patterns of local relatedness (Lukas and Clutton-Brock 2018). In social mammals, complex social relationships are associated with groups that have low relatedness, while members of groups composed of close relatives are more likely to show a diversity of roles (Lukas and Clutton-Brock 2018). While both aspects of social complexity have important implications, it is the measurement of the complexity of social relationships that we attempt to address here.
To have utility, measures of social complexity should be comparable across populations within species, as well as across species, perhaps within some higher taxon. This is challenging. Populations are typically of different sizes, demographics and may use space and interact socially in different ways. Furthermore, they are studied with different protocols and with differing intensities. Ideally, we seek a measure that is as follows: (a) unaffected by network size, so the social complexity calculated from a full social network is similar to that calculated from any substantial random portion of it; (b) little influenced by the addition of distantly connected individuals into the study network; (c) not biased high (suggesting false complexity) by sampling issues; and (d) not biased low (obscuring complexity) by low-intensity sampling. Measures of social complexity can potentially be multidimensional, with different dimensions capturing elements of the concept (e.g., Whitehead 2008; Fischer et al. 2017).
There have been two general perspectives to measuring social complexity using network data. The top-down approach looks at complexity as a network property, using measures such as size, diameter, modularity, dimensional coupling, disparity and computational complexity (Butts 2001; Whitehead 2008). These measures tend to be affected by network delineation, thus causing problems with issues (a) and (b) outlined previously. Indeed, these problems are common to many attempts to develop measures to compare the structure of social networks (Faust 2006).
An alternative, bottom-up, perspective, is to consider social complexity from the perspective of the members of a social network. Hinde (1976) defined social structure as the “nature, quality, and patterning of relationships”. Then, social complexity can be thought of as the complexity of dyadic relationships. If we operationalize relationships using “relationship measures”, such as interaction rates and association indices (Whitehead 2008), these can be used to estimate social complexity. Bergman and Beehner (2015) suggest a simple definition of social complexity as “the number of differentiated relationships that individuals have”. A good example of this relationship-based approach to social complexity, which builds on Bergman and Beehner’s (2015) ideas, is Fischer et al.’s (2017) method. Using detailed observations of affiliative and agonistic interactions, each dyadic relationship is quantified, and, then, these are clustered into one of four relationship classes. Social complexity is quantified using the diversity of relationships experienced by an individual, and individual-level complexities are aggregated into measures of group complexity. While Fischer et al.’s (2017) method is an appealing and rich approach, it depends on the availability of detailed data on direct social interactions (e.g., grooming and aggression), which are often difficult to observe in studies of the social structure of wild animals.
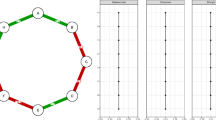
Many studies of social structure employ association indices, estimates of the proportion of time that a dyad is associated (Cairns and Schwager 1987). These association indices are used to infer the structure of social relationships within the population. Association indices (the “simple ratio index”, the “half-weight index”, etc.) are typically calculated as ratios: the number of times that the dyad was observed associating divided by the number of times that they could have been observed associating—a binomial process. Using this attribute of association indices, we introduce a method, which in some respects, parallels that of Fischer et al. (2017), for deriving a measure of social complexity, which we call association complexity, from association indices. We use binomial mixture models on association data to model the distribution of relationships within a population (see Fig. 1). The mixture models represent the associations as belonging to several classes, each with a mean strength of association and rate of occurrence within the population (McNicholas 2016). The mixture modelling finds how many classes are best supported by the data and, then, estimates these parameters. These are then input to a Shannon index of entropy (Shannon and Weaver 1949) to give a measure of diversity among the associations experienced by individuals, which we use to measure complexity.
Illustration of our dyadic concept of association complexity, illustrated for societies of low (a), medium (b) and high (c) complexities. Social networks (left) contain different numbers of relationship types (represented by edge colors), each with a unique distribution of true association indices (centre). We measure complexity as the uncertainty that an association is of a particular relationship type, visualised here as the sum of association indices of each type (right). A more even distribution of sums across more classes of association leads to greater uncertainty, resulting in higher values of S
Here, we first explain the method and, then, test it against simulated data. We explore the effects of sampling rate as well as within-class variability on our estimates of association complexity. Finally, we illustrate the process with real data and discuss potential extensions.
Methods
Binomial mixture models
We assume that each dyad, ij, has a real association index, Rij, that is the actual proportion of time that they are in association and that each Rij belongs to one of K relationship classes, though which class is unknown. So, for instance, there might be some tight “bonded” relationships with Rij = μ1 = 0.75, some pairs of “friends” with Rij = μ2 = 0.20 and some “casual acquaintances” with Rij = μ3 = 0.03.
Then, if the relationship between individual i and individual j is of class k (ij) (the classes, the ks, are labelled 1, 2, 3, …, K; each class with a real association index μk) and there are dij observation occasions, the number of observed associations, xij, is binomially distributed with sample size dij and probability μk(ij). Thus:
We do not know K, the number of classes of relationship, the means for each class, {μk}, or the proportion of relationships in each class, {αk} [Σαk = 1]. However, mixture models allow us to estimate these parameters. Mixture models assume that an observed distribution is a mixture of several unknown distributions and estimate the nature and importance of these different components (McNicholas 2016). In our case, we are trying to dissect a distribution of relationship measures into its components, with each of the components representing a different class of relationship. The parameters [{μk}, {αk}] of the binomial mixture model are estimated using maximum likelihood via an expectation-maximization (EM) algorithm (see the Supplementary material for algorithm details). The number of classes, K, is estimated by fitting a set of candidate models with different values of K and choosing the best one based on criteria, such as the Bayesian Information Criteria (BIC), Akaike Information Criterion (AIC), or the Integrated Completed Likelihood (ICL) (McNicholas 2016). We calculate ICL as BIC + 2E, where E is the entropy of the classification matrix. Thus, ICL penalizes models in which the relationship class of dyads is uncertain.
Quantifying complexity
The mixture models suggest that relationships of class k occur with frequency αk and these dyads associate at a rate of μk (the strength of the association index). Thus, the frequency of associations in the population between two individuals with relationship class k is:
Then, the diversity in association can be expressed by Shannon and Weaver’s (1949) entropy index:
And, this is our proposed measure of association complexity.
This measure has the desirable quality that, in general, social structures with more relationship classes will have a higher value of S. In addition, this measure also quantifies differences in the diversity of associations between social structures with the same number of relationship classes. A society will have higher complexity when the frequency with which classes occur decreases as the strength of association increases. Maximal complexity for a given number of classes is achieved when
As under these conditions, associations of all classes are equally frequent. Deviations from Eq. (4) lead to differences in the frequency of associations of each class, which results in less diversity in association types. Societies with the same value of K can have very different values of S, and difference in values of K will not always reflect differences in S. Stated another way, S indicates the degree of uncertainty in the relationship class of a given association. As an example, consider three hypothetical societies, one with K = 5 and q = {0.2, 0.2, 0.2, 0.2, 0.2}, another with K = 5 and q = {0.9, 0.025, 0.025, 0.025, 0.025}, and a third with K = 2 and q = {0.5, 0.5}. The first two societies have the same number of relationship classes, but in the first, the frequency of associations of each class is the same, and thus, the diversity of associations is extremely high (S = 1.61), while in the second, one class dominates, reducing the association complexity (S = 0.47). Furthermore, while the third society has only two relationship classes, associations of both class are equally likely, leading to an estimate of complexity higher than the second society (S = 0.69). We illustrate the variation in S within and between values of K in our simulations (see subsequent texts).
Testing the method
We used simulated data to test our proposed method. We were particularly interested in which criterion to use for selecting the number of components (AIC, BIC, ICL), as well as how the sampling effort, indicated by the denominator of the association index (dij) might affect estimates of the number of classes of social relationship (K) and association complexity (S). In addition, we sought to more closely simulate real world data by including overdispersion within relationship classes. Overdispersion represents how much more variable observations are than a particular model assumes. In practice, overdispersion from a theoretical distribution could be caused by a variety of behavioural, psychological, environmental or measurement issues. Overdispersion in binomial data is often modelled via beta-binomial distributions. The beta-binomial distribution results from binomial trials in which the probability of success is not constant but follows a beta distribution with shape parameters β1 and β2. In this context, we have found it more useful to consider an alternate parameterization based on the mean, μ = β1/(β1 + β2), and the overdispersion parameter ρ = 1/(β1 + β2 + 1).
The simulations used Poisson and beta-binomial distributions to produce sets of dij and xij, respectively. These simulations were parameterized to reflect the characteristics of real world datasets. We examined six real association datasets (two of which are used as examples, in the subsequent texts) from individually identified wild cetaceans, calculating mean(dij) and estimating overdispersion, ρ, for each. Overdispersion, ρ, was estimated using maximum likelihood assuming the number of components (K), as well as values of {μk} and {αk} are as estimated by the binomial mixture models (using ICL; see subsequent texts). These suggested reasonable ranges of mean(dij) from 15 to 100 and ρ from 0 to 0.01.
We simulated a population of N associating individuals (Ndyad = (N(N − 1)) / 2). We simulated social structure by setting the number of relationship classes, choosing frequencies and distributions of association probabilities for each type, assigning dyads to types and then generating true dyadic association probabilities. We then simulated observational sampling of associations from this social structure. More specifically, in a given simulation run with K relationship classes, we
- 1.
Drew relative αk from a uniform distribution on [0, 1], with the constraint that min (αk) > 0.1/K
- 2.
Drew μk from a uniform distribution on [0, 1], with the constraint that they were at least 0.1 apart
- 3.
Drew ρk from a uniform distribution on [0, 0.015]
- 4.
Assigned k (ij) to dyads with probability αk
- 5.
Generated Rij for each dyad from a beta distribution with mean μk(ij) and overdispersion parameter ρk(ij)
- 6.
Generated dij from a Poisson distribution with mean D
- 7.
Generated xij from a binomial distribution with probability Rij and dij trials
From these simulated social structures, we measured realized association complexity from the k (ij) and Rij and then fit a series of binomial mixture models with K = 1, 2, 3, 4, 5, 6, 7, 8, and 9 to the xij and dij. We chose a best value of K based on BIC, ICL, and AIC and recorded estimates of S based on the models chosen by each of these criteria.
We systematically varied the values of N, K, and D across simulations to test the method under different population sizes, social structures, and sampling effort. We ran 20 simulation runs for every combination of the following parameters: N = 20, 50; K = 1, 2, 3, 4, 5; D = 20, 40, 60, 80, 100.
To examine model performance at estimating S and K, we analysed the mean error in model estimates under different conditions. This gave us a measure of the degree to which our model accurately reflects actual complexity under different conditions, as well as allowing us to examine the model output for bias. We also estimated the correlation between true and estimated values of S for each criterion and under different conditions, to determine the degree to which we can expect the output of the model to reflect differences in complexity between societies.
We also tested our model for sensitivity to systematic increases in overdispersion. Using N = 20, K = 1, 2, 3, 4, 5 and D = 20, 40, 60, 80, 100, we ran simulations in which we defined a common overdispersion parameter ρ for all components. We used ρ = 0.005, 0.01, 0.015, 0.02, running 20 simulations for each combination of parameters. We examined our model for biases introduced by increased overdispersion by analysing the mean error in estimates of S and K in relationship to overdispersion, social structure and sampling.
Illustration using real data
We used two real datasets to illustrate the method. These analyses are illustrative only and are not necessarily optimal analyses of these data. Photoidentification data on 30 northern bottlenose whales (Hyperoodon ampullatus) were collected off Nova Scotia, Canada, between 1988 and 2003, as in Gowans et al. (2001) with some extra data from later years. Photoidentification data on 77 female sperm whales (Physeter macrocephalus) were collected off Dominica, West Indies, between 1984 and 2015, as in Gero et al. (2013a), again, with some extra data. In both studies, sampling periods were days, only individuals identified on more than 10 days were included, association of a dyad was defined as identified within 10 min on the same day, and association indices were calculated using the simple ratio index. For each dataset, we used the binomial mixture model together with the ICL criterion to estimate the number of relationship classes and the characteristics of each, as well as an estimate of association complexity (from Eq. (3)).
Computer code
This work was carried out in parallel and largely independently using the packages R (by MW) and MATLAB (by HW). Functions for using binomial mixture models on association data in both languages are given in the Supplemental material.
Results
Testing the method
As expected, most variation in S in our simulations was driven by differences in the number of relationship classes, as demonstrated by a high correlation between true values of S and K (r = 0.93, Fig. 2). However, when only considering cases in which K > 1 (as when K = 1, S is always 0), the correlation was much lower (r = 0.67), and a significant degree of overlap in values of S between different values of K was apparent (Fig. 2). While the number of relationship classes greatly affects the complexity of associations, the frequency and strength of relationship classes are also an important factor.
Distributions of realized complexity values (S) between societies with different numbers of relationship classes (K). Violin plots represent density estimates and quartiles of true S values for each value of K used. Simulation runs for K = 1 are not plotted as these runs, by definition, have S = 0. Blue points represent the maximum possible entropy for each value of K. Each distribution represents the results of 500 simulation runs
The results of our simulation study largely suggest that ICL is the best criterion to use for these models. The correlation between the estimates of S via ICL and true complexities across all parameters was 0.9, while AIC and BIC had overall correlations of 0.79 and 0.78, respectively. This high correlation for ICL across sampling efforts, network sizes, and social structures indicates that estimates of S based on models chosen via ICL are highly comparable between networks. At low sampling efforts (D < 40), ICL does give estimates of S less correlated with true complexities than AIC or BIC, but it rapidly tends towards a perfect correlation with increased sampling effort. In contrast, the correlations between true and estimated complexities obtained by AIC and BIC do not increase with sampling effort and are consistently below 0.9 (Fig. 3, left).
Correlation between real and estimated S (left) and mean error in estimates of S (right) for each criterion under different levels of sampling effort (expressed as mean denominator, D) and network sizes (in number of individuals, N). Each data point is based on 250 simulation runs (50 runs for each value of K). Dotted black line indicates a mean error of 0
AIC and BIC were both likely to overestimate the complexity of a social structure, and this overestimation was exacerbated by increased sampling effort. In contrast, the estimates obtained by ICL are downward biased at low sampling rates, but the bias decreases as sampling effort increases. This indicates that ICL estimates are unlikely to be overestimates of true complexity, but large amounts of data (D > 80) are likely needed to ensure accurate estimates. However, even at low sampling rates, the bias is less than 0.5 (Fig. 3, right).
In addition, both AIC and BIC provide estimates that are sensitive to network size in our simulations, with larger networks having added positive bias. In contrast, ICL did not give estimates biased by network size (Fig. 3) and, thus, provide an estimate of complexity that is comparable between social networks of different sizes and levels of completeness (a reasonable, roughly random subset of a larger network should provide a similar estimate as the full network).
ICL was prone to underestimating both S and K at low sampling rates. This tendency was exacerbated by social structures with more relationship classes. This bias was relieved with increased sampling effort. In addition, ICL rarely found multiple relationship classes in social structures in which there was only one class of dyad (Fig. 4). Therefore, while we suggest the use of ICL to choose the number of components in these models, as it gives good estimates that are comparable between networks, we caution that these estimates will likely be underestimated with low sampling intensity, particularly for complex social structures.
Relationship between input value of K and error in estimates of S and K obtained from models chosen via ICL. Colors indicate simulated sampling effort (as expressed by mean denominator of association indices, D). Results are presented based on runs with N = 20, and each data point represents the mean of 50 simulation runs. Dotted black line indicates a mean error of 0
All criteria were somewhat sensitive to systematic increases in overdispersion. High levels of overdispersion led to overestimates of complexity, particularly under high sampling intensity. However, ICL was far less sensitive to overdispersion than AIC or BIC. At values of ρ < 0.015, ICL converged towards zero bias as sampling effort increased towards D = 100, and even at ρ = 0.015, upward bias at high sampling intensity was small. At ρ = 0.02, upward bias at high sampling intensities became more pronounced (Fig. 5).
Illustration using real data
The distributions of simple ratio association indices for the northern bottlenose whale and sperm whale datasets are shown in Fig. 5. Mixture models suggested 2 relationship classes for the northern bottlenose whales with an association complexity of S = 0.69 and 3 relationship classes for the sperm whales with an association complexity of S = 0.91. The mean denominators of the association indices and estimates of overdispersion were D = 34.6 and ρ = 0.010 for the northern bottlenose whales and D = 59.9 and ρ = 0.007 for the sperm whales. Using the simulation data in Fig. 4, these suggest that our model estimates may have small (< 0.2) downward biases.
Figure 6 shows the estimated distribution of real association indices from the binomial mixture models and estimates of overdispersion. While they roughly match the distribution of measured association indices, the matching is not too good, but it is must be remembered that the measured association indices include sampling error while the estimated real association indices do not.
Both species have a preponderance of extremely low association relationships (μ1 = 0.017 and α1 = 0.88 for the northern bottlenose whales; μ1 = 0.002 and α1 = 0.90 for the sperm whales), as well as some low association relationships (μ2 = 0.125 and α2 = 0.12 for the northern bottlenose whales; μ2 = 0.072 and α2 = 0.07 for the sperm whales). The sperm whales additionally have a much smaller class of fairly strong association relationships (μ3 = 0.252 and α3 = 0.03). The latter correspond to relationships within social units (Gero et al. 2013a).
Discussion
We have presented a method for quantifying the complexity of association networks based on dyadic sighting histories. We use binomial mixture models to estimate the number of different classes of relationship and the association frequencies of each class and take the diversity of these frequencies as our measure of association complexity. Our results show that this approach can generally be used to effectively model the dyadic associations and measure network complexity and is comparable between networks.
Hinde (1976) defined social structure as the “nature, quality, and patterning of relationships”. Ideally, we would measure complexity from all three of these elements. However, it is well-known that measures of the global patterning of relationships—such as metrics from network analysis—are not comparable between networks, due to the dependency of these measures on network size and density (Faust 2006; Rito et al. 2010; van Wijk et al. 2010). This is a significant problem for the field of animal social networks because it makes the comparative approach difficult. Our method instead examines social complexity through the nature and quality of dyadic relationships—providing a bottom-up measure of complexity that can be fairly compared between association networks. Our method can therefore be used with a comparative approach to examine drivers of social complexity across populations, species and potentially taxa.
A previous approach to measuring dyadic complexity (Fischer et al. 2017) is a promising way forward for many systems, but it is not appropriate for association data, because it requires classes of interaction to be known and pre-defined in the complexity measure. The researcher needs data more detailed than just who was with whom (associations) and on whether an interaction is of the class aggressive or the class affiliative. Our approach instead seeks to automatically identify different classes of dyad based on the patterns of associations. The same limitations that apply to any analysis using association indices apply to our method. Since all that is being measured and modelled is the proportion of time individuals spend together, the nuances of social relationships are perhaps not captured by these measures. For example, our method would not be able to distinguish between two relationship classes that associate with the same probability but interact in different ways while associated. We suggest that our model will be a useful comparative tool when the collection of detailed interaction data is impractical, such as in studies of wild cetaceans.
Our complexity measure is unaffected by network size; since our measure is based on dyads, the association complexity of a reasonably well-sampled social network will be similar to that of the full network. Our measure is also fairly robust to the existence of individuals that are distantly connected to the network and thus observed infrequently. Although our method rarely estimates a higher level of complexity than that of the true network, low-intensity sampling biases it towards artificially low estimates of complexity. It is a common feature of social network analysis that low-intensity sampling produces metrics that are unreliable (Whitehead 2008; Franks et al. 2010; Farine and Whitehead 2015), and we, therefore, suggest that caution is taken when interpreting results from this model on sparsely sampled data.
Because the complexity measure is partly based on unevenness of dyadic weights, we might expect a network sampled with the gambit of the group to have a higher level of complexity than a network sampled by observing pairwise associations (e.g., by focal sampling). This is because there will be more casual acquaintances in the network as an artefact of the gambit sampling method. For example, both individuals A and B might only be observed together because they are both associating with individual C. Thus, when adopting a comparative approach, differences in sampling protocol will need to be considered.
Finally, the driver of association complexity needs to be considered for each social system, because complex social structures can arise through a number of mechanisms. Complex social structures, such as multilevel societies, can arise from cognitively demanding behavioural processes, such as cultural transmission (Cantor et al. 2015). However, complexity can also be driven by simple differences between individuals in their social behaviours (Firth et al. 2017). Furthermore, there is increasing recognition of the role that features of the physical environment play in shaping social structures (He et al. 2019, Topical collection on Social complexity). Therefore, it could be that the social decisions of individuals do not produce a complex network, but instead social complexity is driven by patterns of space use or the complexity of the environment (Titcomb et al. 2015; Leu et al. 2016). Complex patterns of overlapping space use could lead to higher estimates of social complexity with our method. It is therefore important that our proposed metric not be interpreted as a measure of the complexity of individuals’ social decision-making but rather as a feature of the social structure of the population.
If our measure of association complexity is to be widely used, it needs some measure of confidence. We suggest the temporal jackknife, in which different temporal segments of data are omitted in turn. This method is appropriate with behavioural association data when the nonparametric bootstrap cannot be used (as randomizing identities produces self-associations) (Whitehead 2008). Additionally, it would be helpful to give analytic estimates of the bias due to sampling rates and overdispersion that are indicated by our sensitivity analyses. There also could be more robust measures of association complexity from mixture model data that perform better than the Shannon index, but we have not yet found any.
The method that we have proposed could be varied or extended in several potentially productive ways. Using the same dataset, two or more measures of association could be defined, based on different behavioural states or ways of associating (e.g., Gero et al. 2005, 2013b). These, then, constitute multivariate relationship measures, which could be clustered using multivariate mixture models (McNicholas 2016). To obtain our univariate measure of association complexity, using Eqs. (2) and (3), we need some way of compounding the now vector-valued centroids of the clusters (μs), perhaps using principal components analysis. However, we could also calculate separate measures of complexity for each association measure, so that, for instance, complexity could be compared between behavioural states or modes of communication. Our association complexity measure(s) could also be used in parallel with other network or relationship measures, such as modularity (Newman 2006), to give a more nuanced comparison between social networks.
Many social network data are in the form of interaction rates (Farine and Whitehead 2015). Poisson mixture models would be appropriate in these cases, perhaps with offset variables indicating effort. These interaction rate data could be combined with each other, or with association data, in a multivariate mixture analysis. Offset variables may be useful more generally. For instance, generalized affiliation indices are the residuals from a regression of the measures of association or interaction on structural predictor variables, such as gregariousness or spatiotemporal overlap (Whitehead and James 2015). Inputting generalized affiliation indices into mixture models, either directly into Gaussian mixtures or as offsets in binomial or Poisson mixtures, could control for use of space and other confounds.
We have attached R and Matlab code for deriving association complexity using mixture models, and the method will also be incorporated in the next release of SOCPROG, a package for analysing animal social structures using individual identification data (Whitehead 2009).
References
Bergman TJ, Beehner JC (2015) Measuring social complexity. Anim Behav 103:203–209. https://doi.org/10.1016/j.anbehav.2015.02.018
Blumenstein DT, Armitage KB (1998) Life history consequences of social complexity: a comparative study of ground-dwelling sciurids. Behav Ecol 9:8–19
Butts CT (2001) The complexity of social networks: theoretical and empirical findings. Soc Networks 23:31–72. https://doi.org/10.1016/S0378-8733(01)00030-2
Cairns SJ, Schwager SJ (1987) A comparison of association indices. Anim Behav 35:1454–1469
Cantor M, Shoemaker LG, Cabral RB, Flores CO, Varga M, Whitehead H (2015) Multilevel animal societies can emerge from cultural transmission. Nat Commun 6:8091
Farine DR, Whitehead H (2015) Constructing, conducting and interpreting animal social network analysis. J Anim Ecol 84:1144–1163
Faust K (2006) Comparing social networks: size, density, and local structure. Metodoloski Zvezki 3:185
Firth JA, Sheldon BC, Brent LJN (2017) Indirectly connected: simple social differences can explain the causes and apparent consequences of complex social network positions. Proc R Soc B 284:20171939. https://doi.org/10.1098/rspb.2017.1939
Fischer J, Farnworth MS, Sennhenn-Reulen H, Hammerschmidt K (2017) Quantifying social complexity. Anim Behav 130:57–66. https://doi.org/10.1016/j.anbehav.2017.06.003
Franks DW, Ruxton GD, James R (2010) Sampling animal association networks with the gambit of the group. Behav Ecol Sociobiol 64:493–503
Freeberg TM, Dunbar RI, Ord TJ (2012) Social complexity as a proximate and ultimate factor in communicative complexity. Philos Trans R Soc B 367:1785–1801. https://doi.org/10.1098/rstb.2011.0213
Gero S, Bejder L, Whitehead H, Mann J, Connor RC (2005) Behaviourally specific preferred associations in bottlenose dolphins, Tursiops sp. Can J Zool 83:1566–1573
Gero S, Milligan M, Rinaldi C, Francis P, Gordon J, Carlson C, Steffen A, Tyack P, Evans P, Whitehead H (2013a) Behavior and social structure of the sperm whales of Dominica, West Indies. Mar Mammal Sci 30:905–922
Gero S, Gordon J, Whitehead H (2013b) Calves as social hubs: dynamics of the social network within sperm whale units. Proc R Soc B 280:20131113
Gowans S, Whitehead H, Hooker SK (2001) Social organization in northern bottlenose whales (Hyperoodon ampullatus): not driven by deep water foraging? Anim Behav 62:369–377
Groenewoud F, Frommen JG, Josi D, Tanaka H, Jungwirth A, Taborsky M (2016) Predation risk drives social complexity in cooperative breeders. Proc Natl Acad Sci USA 113:4104–4109. https://doi.org/10.1073/pnas.1524178113
He P, Malonado-Chaparro A, Farine DR (2019) The role of habitat configuration in shaping social structure: a gap in studies of animal social complexity. Behav Ecol Sociobiol this issue. https://doi.org/10.1007/s00265-018-2602-7
Hinde RA (1976) Interactions, relationships and social structure. Man 11:1–17
Kappeler PM (2019) A framework for studying social complexity. Behav Ecol Sociobiol this issue. https://doi.org/10.1007/s00265-018-2601-8
Kwak S, Joo W, Youm Y, Chey J (2018) Social brain volume is associated with in-degree social network size among older adults. Proc R Soc B 285:20172708
Leu ST, Farine DR, Wey TW, Sih A, Bull CM (2016) Environment modulates population social structure: experimental evidence from replicated social networks of wild lizards. Anim Behav 111:23–31
Lukas D, Clutton-Brock T (2018) Social complexity and kinship in animal societies. Ecol Lett (published online, doi:https://doi.org/10.1111/ele.13079)
McNicholas PD (2016) Model-based clustering. J Classif 33:331–373
Newman MEJ (2006) Modularity and community structure in networks. Proc Natl Acad Sci USA 103:8577–8582
Rito T, Wang Z, Deane CM, Reinert G (2010) How threshold behaviour affects the use of subgraphs for network comparison. Bioinformatics 26:i611–i617
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Titcomb EM, O'Corry-Crowe G, Hartel EF, Mazzoil MS (2015) Social communities and spatiotemporal dynamics of association patterns in estuarine bottlenose dolphins. Mar Mammal Sci 31:1314–1337
van Wijk BC, Stam CJ, Daffertshofer A (2010) Comparing brain networks of different size and connectivity density using graph theory. PLoS One 5:e13701
Whitehead H (2008) Analyzing animal societies: quantitative methods for vertebrate social analysis. Chicago University Press, Chicago
Whitehead H (2009) SOCPROG programs: analysing animal social structures. Behav Ecol Sociobiol 63:765–778
Whitehead H, James R (2015) Generalized affiliation indices extract affiliations from social network data. Methods Ecol Evol 6:836–844
Acknowledgments
Thanks to Shane Gero for the Dominica sperm whale data and to two anonymous reviewers for constructive comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by T. Clutton-Brock
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a contribution to the Topical Collection Social complexity: patterns, processes, and evolution – Guest Editors: Peter Kappeler, Susanne Shultz, Tim Clutton-Brock, and Dieter Lukas
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Weiss, M.N., Franks, D.W., Croft, D.P. et al. Measuring the complexity of social associations using mixture models. Behav Ecol Sociobiol 73, 8 (2019). https://doi.org/10.1007/s00265-018-2603-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00265-018-2603-6