Abstract
Objective
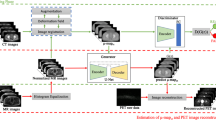
Quantitative PET/MR imaging is challenged by the accuracy of synthetic CT (sCT) generation from MR images. Deep learning-based algorithms have recently gained momentum for a number of medical image analysis applications. In this work, a novel sCT generation algorithm based on deep learning adversarial semantic structure (DL-AdvSS) is proposed for MRI-guided attenuation correction in brain PET/MRI.
Materials and methods
The proposed DL-AdvSS algorithm exploits the ASS learning framework to constrain the synthetic CT generation process to comply with the extracted structural features from CT images. The proposed technique was evaluated through comparison to an atlas-based sCT generation method (Atlas), previously developed for MRI-only or PET/MRI-guided radiation planning. Moreover, the commercial segmentation-based approach (Segm) implemented on the Philips TF PET/MRI system was included in the evaluation. Clinical brain studies of 40 patients who underwent PET/CT and MR imaging were used for the evaluation of the proposed method under a two-fold cross validation scheme.
Results
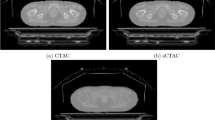
The accuracy of cortical bone extraction and CT value estimation were investigated for the three different methods. Atlas and DL-AdvSS exhibited similar cortical bone extraction accuracy resulting in a Dice coefficient of 0.78 ± 0.07 and 0.77 ± 0.07, respectively. Likewise, DL-AdvSS and Atlas techniques performed similarly in terms of CT value estimation in the cortical bone region where a mean error (ME) of less than −11 HU was obtained. The Segm approach led to a ME of −1025 HU. Furthermore, the quantitative analysis of corresponding PET images using the three approaches assuming the CT-based attenuation corrected PET (PETCTAC) as reference demonstrated comparative performance of DL-AdvSS and Atlas techniques with a mean standardized uptake value (SUV) bias less than 4% in 63 brain regions. In addition, less that 2% SUV bias was observed in the cortical bone when using Atlas and DL-AdvSS approaches. However, Segm resulted in 14.7 ± 8.9% SUV underestimation in the cortical bone.
Conclusion
The proposed DL-AdvSS approach demonstrated competitive performance with respect to the state-of-the-art atlas-based technique achieving clinically tolerable errors, thus outperforming the commercial segmentation approach used in the clinic.







Similar content being viewed by others
References
Zaidi H, Becker M. The promise of hybrid PET/MRI: technical advances and clinical applications. IEEE Signal Process Mag. 2016;33:67–85.
Mehranian A, Arabi H, Zaidi H. Vision 20/20: magnetic resonance imaging-guided attenuation correction in PET/MRI: challenges, solutions, and opportunities. Med Phys. 2016;43:1130–55.
Erlandsson K, Dickson J, Arridge S, Atkinson D, Ourselin S, Hutton BF. MR imaging-guided partial volume correction of PET data in PET/MR imaging. PET Clinics. 2016;11:161–77.
Catana C. Motion correction options in PET/MRI. Semin Nucl Med. 2015;45:212–23.
Mehranian A, Belzunce M, Prieto C, Hammers A, Reader A. Synergistic PET and SENSE MR image reconstruction using joint sparsity regularization. IEEE Trans Med Imaging. 2018;37:20–34.
Gong K, Cheng-Liao J, Wang G, Chen KT, Catana C, Qi J. Direct patlak reconstruction from dynamic PET data using the kernel method with MRI information based on structural similarity. IEEE Trans Med Imaging. 2018;37:955–65.
Zaidi H, Montandon M-L, Slosman DO. Magnetic resonance imaging-guided attenuation and scatter corrections in three-dimensional brain positron emission tomography. Med Phys. 2003;30:937–48.
Martinez-Moller A, Souvatzoglou M, Delso G, Bundschuh RA, Chefd’hotel C, Ziegler SI, et al. Tissue classification as a potential approach for attenuation correction in whole-body PET/MRI: evaluation with PET/CT data. J Nucl Med. 2009;50:520–6.
Keereman V, Fierens Y, Broux T, De Deene Y, Lonneux M, Vandenberghe S. MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J Nucl Med. 2010;51:812–8.
Arabi H, Rager O, Alem A, Varoquaux A, Becker M, Zaidi H. Clinical assessment of MR-guided 3-class and 4-class attenuation correction in PET/MR. Mol Imaging Biol. 2015;17:264–76.
Wollenweber SD, Ambwani S, Delso G, Lonn AHR, Mullick R, Wiesinger F, et al. Evaluation of an atlas-based PET head attenuation correction using PET/CT & MR patient data. IEEE Trans Nucl Sci. 2013;60:3383–90.
Arabi H, Zaidi H. One registration multi-atlas-based pseudo-CT generation for attenuation correction in PET/MRI. Eur J Nucl Med Mol Imaging. 2016;43:2021–35.
Burgos N, Cardoso MJ, Guerreiro F, Veiga C, Modat M, McClelland J, et al. Robust CT synthesis for radiotherapy planning: application to the head and neck region. In: International conference on medical image computing and computer-assisted intervention: Springer; 2015. p. 476–84.
Defrise M, Rezaei A, Nuyts J. Time-of-flight PET data determine the attenuation sinogram up to a constant. Phys Med Biol. 2012;57:885–99.
Mehranian A, Zaidi H, Reader AJ. MR-guided joint reconstruction of activity and attenuation in brain PET-MR. NeuroImage. 2017;162:276–88.
Rezaei A, Deroose CM, Vahle T, Boada F, Nuyts J. Joint reconstruction of activity and attenuation in fime-of-flight PET: a quantitative analysis. J Nucl Med. 2018;59:1624–9.
Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, et al. Estimating CT image from MRI data using structured random forest and auto-context model. IEEE Trans Med Imaging. 2016;35:174–83.
Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–19.
Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Zero-echo-time and Dixon deep pseudo-CT (ZeDD CT): direct generation of pseudo-CT images for pelvic PET/MRI attenuation correction using deep convolutional neural networks with multiparametric MRI. J Nucl Med. 2018;59:852–8.
Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, et al. Medical image synthesis with context-aware generative adversarial network. In: Medical image computing and computer-assisted intervention − MICCAI 2017, Quebec, Canada; Springer, 2017. pp 417–25.
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286:676–84.
Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35:1153–9.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Emami H, Dong M, Nejad-Davarani SP, Glide-Hurst CK. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. 2018;45:3627–36.
Dinkla AM, Wolterink JM, Maspero M, Savenije MHF, Verhoeff JJC, Seravalli E, et al. MR-only brain radiation therapy: dosimetric evaluation of synthetic CTs generated by a dilated convolutional neural network. Int J Radiat Oncol Biol Phys. 2018;102:801–12.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9.
Chen S, Qin A, Zhou D, Yan D. Technical note: U-net-generated synthetic CT images for magnetic resonance imaging-only prostate intensity-modulated radiation therapy treatment planning. Med Phys. 2018;45:5659–65.
Xiang L, Wang Q, Nie D, Zhang L, Jin X, Qiao Y, et al. Deep embedding convolutional neural network for synthesizing CT image from T1-weighted MR image. Med Image Anal. 2018;47:31–44.
Arabi H, Koutsouvelis N, Rouzaud M, Miralbell R, Zaidi H. Atlas-guided generation of pseudo-CT images for MRI-only and hybrid PET–MRI-guided radiotherapy treatment planning. Phys Med Biol. 2016;61:6531–52.
Arabi H, Dowling JA, Burgos N, Han X, Greer PB, Koutsouvelis N, et al. Comparative study of algorithms for synthetic CT generation from MRI: consequences for MRI-guided radiation planning in the pelvic region. Med Phys. 2018;45:5218–33.
Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd international conference on machine learning, Lille, France, vol. 37; 2015. pp 448–56.
Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90.
Zaidi H, Ojha N, Morich M, Griesmer J, Hu Z, Maniawski P, et al. Design and performance evaluation of a whole-body ingenuity TF PET-MRI system. Phys Med Biol. 2011;56:3091–106.
Zaidi H, Montandon M-L, Meikle S. Strategies for attenuation compensation in neurological PET studies. Neuroimage. 2007;34:518–41.
Mehranian A, Arabi H, Zaidi H. Quantitative analysis of MRI-guided attenuation correction techniques in time-of-flight brain PET/MRI. NeuroImage. 2016;130:123–33.
Chartsias A, Joyce T, Dharmakumar R, Tsaftaris SA. Adversarial image synthesis for unpaired nulti-modal cardiac data. In: International workshop on simulation and synthesis in medical imaging, SASHIMI; 2017. pp 3–13.
Huo Y, Xu Z, Bao S, Assad A, Abramson RG, Landman BA. Adversarial synthesis learning enables segmentation without target modality ground truth. In: IEEE 15th International Symposium on Biomedical Imaging (ISBI); 2018. pp 1217–20.
Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. IEEE international conference on computer vision. 2017. pp. 2223-2232.
Su KH, Hu L, Stehning C, Helle M, Qian P, Thompson CL, et al. Generation of brain pseudo-CTs using an undersampled, single-acquisition UTE-mDixon pulse sequence and unsupervised clustering. Med Phys. 2015;42:4974–86.
Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol. 2018;63:125011.
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for 18 F-FDG PET attenuation correction. EJNMMI Phys. 2018;5:24.
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging–based attenuation correction for PET/MR imaging. Radiology. 2017;286:676–84.
Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol. 2018;63(12):125011.
Spuhler KD, Gardus J, Gao Y, DeLorenzo C, Parsey R, Huang C. Synthesis of patient-specific transmission image for PET attenuation correction for PET/MR imaging of the brain using a convolutional neural network. J Nucl Med. 2019;60:555–60.
Acknowledgments
This work was supported by the Swiss National Science Foundation under grant SNFN 320030_176052 and the Swiss Cancer Research Foundation under Grant KFS-3855-02-2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
None of the authors have affiliations that present financial or non-financial competing interests for this work.
Research involving human participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
ESM 1
(DOCX 717 kb)
Rights and permissions
About this article
Cite this article
Arabi, H., Zeng, G., Zheng, G. et al. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur J Nucl Med Mol Imaging 46, 2746–2759 (2019). https://doi.org/10.1007/s00259-019-04380-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04380-x