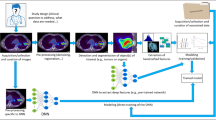
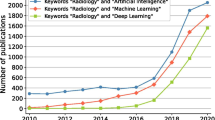
Abstract
Techniques from the field of artificial intelligence, and more specifically machine (deep) learning methods, have been core components of most recent developments in the field of medical imaging. They are already being exploited or are being considered to tackle most tasks, including image reconstruction, processing (denoising, segmentation), analysis and predictive modelling. In this review we introduce and define these key concepts and discuss how the techniques from this field can be applied to nuclear medicine imaging applications with a particular focus on radio(geno)mics.


Similar content being viewed by others
References
Beyer T, Townsend DW, Brun T, Kinahan PE, Charron M, Roddy R, et al. A combined PET/CT scanner for clinical oncology. J Nucl Med. 2000;41:1369–79.
Lecoq P. Pushing the limits in time-of-flight PET imaging. IEEE Trans Radiat Plasma Med Sci. 2017;1(6):473–85.
Berg E, Cherry SR. Innovations in instrumentation for positron emission tomography. Semin Nucl Med. 2018;48:311–31.
Aerts H. Radiomics: there is more than meets the eye in medical imaging. Plenary presentation from SPIE Medical Imaging 2016: computer-aided diagnosis. https://doi.org/10.1117/12.2214251.
Hatt M, Tixier F, Visvikis D, Cheze Le Rest C. Radiomics in PET/CT: more than meets the eye? J Nucl Med. 2017;58:365–6.
Moor J. The Dartmouth College Artificial Intelligence Conference: the next fifty years. Al Mag. 2006;27:87–91.
Kaplan A, Haenlein M. Siri, Siri, in my hand: Who’s the fairest in the land? On the interpretations, illustrations, and implications of artificial intelligence. Bus Horiz. 2019;62:15–25.
Maloof M. Artificial intelligence: an introduction. Washington DC: Georgetown University. 2015. http://people.cs.georgetown.edu/~maloof/cosc270.f17/cosc270-intro-handout.pdf.
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484–9.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10.
Palmer WJ. Artificial intelligence in radiology: friend or foe? Diagnostic Imaging. 2019. http://www.diagnosticimaging.com/di-executive/artificial-intelligence-radiology-friend-or-foe.
Duda RO, Hart PE, Stork DG. Pattern classification. New York: Wiley; 2012.
Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge, MA: MIT Press; 2012.
Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–82.
Phuong TM, Lin Z, Altman RB. Choosing SNPs using feature selection. Proceedings of the 2005 IEEE Computational Systems Bioinformatics Conference. 2005; p. 301–9.
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117.
Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems, vol. 1; 2012. p. 1097–105.
Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Berlin: Springer; 2013. p. 411–8.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115:211–52.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ. Quantitative imaging of cancer in the postgenomic era: radio(geno)mics, deep learning, and habitats. Cancer. 2018;124:4633–49.
Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. 2013. p. 8609–13.
Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops: CVPRW 2014. Washington, DC, USA: IEEE Computer Society; 2014. p. 512–519. https://doi.org/10.1109/CVPRW.2014.131.
Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–59.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–98.
Le Pogam A, Hanzouli H, Hatt M, Cheze Le Rest C, Visvikis D. Denoising of PET images by combining wavelets and curvelets for improved preservation of resolution and quantitation. Med Image Anal. 2013;17:877–91.
Gong K, Guan J, Liu C, Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):153–61.
Le Pogam A, Hatt M, Descourt P, Boussion N, Tsoumpas C, Turkheimer FE, et al. Evaluation of a 3D local multiresolution algorithm for the correction of partial volume effects in positron emission tomography. Med Phys. 2011;38:4920–3.
Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-net convolutional neural network study. PLoS One. 2018;13:e0195798.
Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177–95.
Hatt M, Lee JA, Schmidtlein CR, Naqa IE, Caldwell C, De Bernardi E, et al. Classification and evaluation strategies of auto-segmentation approaches for PET: report of AAPM task group no. 211. Med Phys. 2017;44:e1–42.
Zhao X, Li L, Lu W, Tan S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys Med Biol. 2018;64:015011.
Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol. 2010;65:517–21.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6.
Bai HX, Lee AM, Yang L, Zhang P, Davatzikos C, Maris JM, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89:20151030.
Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–41.
Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–80.
Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
Hatt M, Parmar C, Qi J, Naqa IE. Machine (deep) learning methods for image processing and radiomics. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):104–8.
Hong X, Zan Y, Weng F, Tao W, Peng Q, Huang Q. Enhancing the image quality via transferred deep residual learning of coarse PET sinograms. IEEE Trans Med Imaging. 2018;37:2322–32.
Berg E, Cherry SR. Using convolutional neural networks to estimate time-of-flight from PET detector waveforms. Phys Med Biol. 2018;63:02LT01.
Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2018;38:675–85.
Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Trans Med Imaging. 2018;37:1478–87.
Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. In: Deep Learning and Data Labeling for Medical Applications – First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, held in conjunction with MICCAI 2016, Athens, Greece, 21 October 2016; 2016. p. 170–8.
Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med. 2019;60(3):429–35.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019. https://doi.org/10.2967/jnumed.118.219493.
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286(2):676–84.
Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Zero-echo-time and Dixon deep pseudo-CT (ZeDD CT): direct generation of pseudo-CT images for pelvic PET/MRI attenuation correction using deep convolutional neural networks with multiparametric MRI. J Nucl Med. 2018;59:852–8.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9.
Kaplan S, Zhu Y-M. Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging. 2018. https://doi.org/10.1007/s10278-018-0150-3.
Perk T, Bradshaw T, Chen S, Im H-J, Cho S, Perlman S, et al. Automated classification of benign and malignant lesions in 18F-NaF PET/CT images using machine learning. Phys Med Biol. 2018;63:225019.
Zhong Z, Kim Y, Plichta K, Allen BG, Zhou L, Buatti J, et al. Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med Phys. 2018;46(2):619–33.
Guo Z, Li X, Huang H, Guo N, Li Q. Deep learning-based image segmentation on multi-modal medical imaging. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):162–9.
Schwyzer M, Ferraro DA, Muehlematter UJ, Curioni-Fontecedro A, Huellner MW, von Schulthess GK, et al. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks – initial results. Lung Cancer. 2018;126:170–3.
Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zöphel K, et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7:13206.
Deist TM, Dankers FJWM, Valdes G, Wijsman R, Hsu I-C, Oberije C, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med Phys. 2018;45:3449–59.
Upadhaya T, Vallières M, Chatterjee A, Lucia F, Bonaffini PA, Masson I, et al. Comparison of radiomics models built through machine learning in a multicentric context with independent testing: identical data, similar algorithms, different methodologies. IEEE Trans Radiat Plasma Med Sci. 2018;3(2):192–200.
Wu J, Lian C, Ruan S, Mazur TR, Mutic S, Anastasio MA, et al. Treatment outcome prediction for cancer patients based on radiomics and belief function theory. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):216–24.
Klyuzhin IS, Fu JF, Shenkov N, Rahmim A, Sossi V. Use of generative disease models for analysis and selection of radiomic features in PET. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):178–91.
Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Bist V, et al. An empirical approach for avoiding false discoveries when applying high-dimensional radiomics to small datasets. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):201–9.
Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Saif S, et al. Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):210–5.
Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711.
Paul R, Hall L, Goldgof D, Schabath M, Gillies R. Predicting nodule malignancy using a CNN ensemble approach. Proc Int Jt Conf Neural Netw. 2018. https://doi.org/10.1109/IJCNN.2018.8489345.
Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290:290–7.
Bibault J-E, Giraud P, Housset M, Durdux C, Taieb J, Berger A, et al. Deep learning and radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611.
Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7:5467.
Cha KH, Hadjiiski L, Chan H-P, Weizer AZ, Alva A, Cohan RH, et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep. 2017;7:8738.
Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, et al. Convolutional neural network using a breast MRI tumor dataset can predict oncotype Dx recurrence score. J Magn Reson Imaging. 2019;49:518–24.
Ypsilantis P-P, Siddique M, Sohn H-M, Davies A, Cook G, Goh V, et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS One. 2015;10:e0137036.
Amyar A, Ruan S, Gardin I, Chatelain C, Decazes P, Modzelewski R. 3D RPET-NET: development of a 3D PET imaging convolutional neural network for radiomics analysis and outcome prediction. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):225–31.
Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, et al. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2019;290:456–64.
Mabrouk R, Chikhaoui B, Bentabet L. Machine learning based classification using clinical and DaTSCAN SPECT imaging features: a study on Parkinson’s disease and SWEDD. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):170–7.
Wong KCL, Syeda-Mahmood T, Moradi M. Building medical image classifiers with very limited data using segmentation networks. Med Image Anal. 2018;49:105–16.
Yosinski J, Clune J, Nguyen A, Fuchs T, Lipson H. Understanding neural networks through deep visualization. arXiv:1506.06579 [cs.CV]. http://arxiv.org/abs/1506.06579.
Ford R, Price W. Privacy and accountability in black-box medicine. Mich Telecommun Technol Law Rev. 2016;23:1–43.
Vallières M, Zwanenburg A, Badic B, Cheze-Le Rest C, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. 2018;59(2):189–93.
Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv:1612.07003 [cs.CV]. http://arxiv.org/abs/1612.07003.
Hatt M, Vallières M, Visvikis D, Zwanenburg A. IBSI: an international community radiomics standardization initiative. J Nucl Med. 2018;59:287.
Lucia F, Visvikis D, Desseroit M-C, Miranda O, Malhaire J-P, Robin P, et al. External validation of a combined PET and MRI radiomics model for prediction of distance recurrence and locoregional control in locally advanced cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2019;46(4):864–77.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
N/A.
Conflict of interest
None.
Ethical approval
This is a review article for which ethical approval is not required.
Informed consent
This is a review article for which informed consent is not relevant.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Rights and permissions
About this article
Cite this article
Visvikis, D., Cheze Le Rest, C., Jaouen, V. et al. Artificial intelligence, machine (deep) learning and radio(geno)mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging 46, 2630–2637 (2019). https://doi.org/10.1007/s00259-019-04373-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04373-w