Abstract
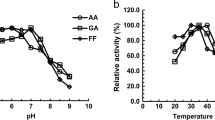
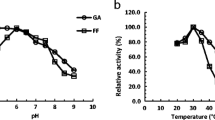
At least 24 aldehyde reductases from Saccharomyces cerevisiae have been characterized and most function in in situ detoxification of lignocellulosic aldehyde inhibitors, but none is classified into the polyol dehydrogenase (PDH) subfamily of the medium-chain dehydrogenase/reductase (MDR) superfamily. This study confirmed that two (2R,3R)-2,3-butanediol dehydrogenases (BDHs) from industrial (denoted Y)/laboratory (denoted B) strains of S. cerevisiae, Bdh1p(Y)/Bdh1p(B) and Bdh2p(Y)/Bdh2p(B), were members of the PDH subfamily with an NAD(P)H binding domain and a catalytic zinc binding domain, and exhibited reductive activities towards lignocellulosic aldehyde inhibitors, such as acetaldehyde, glycolaldehyde, and furfural. Especially, the highest enzyme activity towards acetaldehyde by Bdh2p(Y) was 117.95 U/mg with cofactor nicotinamide adenine dinucleotide reduced (NADH). Based on the comparative kinetic property analysis, Bdh2p(Y)/Bdh2p(B) possessed higher specific activity, substrate affinity, and catalytic efficiency towards glycolaldehyde than Bdh1p(Y)/Bdh1p(B). This was speculated to be related to their 49% sequence differences and five nonsynonymous substitutions (Ser41Thr, Glu173Gln, Ile270Leu, Ile316Met, and Gly317Cys) occurred in their conserved NAD(P)H binding domains. Compared with BDHs from a laboratory strain, Bdh1p(Y) and Bdh2p(Y) from an industrial strain displayed five nonsynonymous mutations (Thr12, Asn61, Glu168, Val222, and Ala235) and three nonsynonymous mutations (Ala34, Ile96, and Ala369), respectively. From a first analysis with selected aldehydes, their reductase activities were different from BDHs of laboratory strain, and their catalytic efficiency was higher towards glycolaldehyde and lower towards acetaldehyde. Comparative investigation of kinetic properties of BDHs from S. cerevisiae as aldehyde reductases provides a guideline for their practical applications in in situ detoxification of aldehyde inhibitors during lignocellulose bioconversion.
Key Points
• Two yeast BDHs have enzyme activities for reduction of aldehydes.
• Overexpression of BDHs slightly improves yeast tolerance to acetaldehyde and glycolaldehyde.
• Bdh1p and Bdh2p differ in enzyme kinetic properties.
• BDHs from strains with different genetic backgrounds differ in enzyme kinetic properties.







Similar content being viewed by others
References
Adrover M, Vilanova B, Munoz F, Donoso J (2008) Kinetic study of the reaction of glycolaldehyde with two glycation target models. Ann N Y Acad Sci 1126:235–240. https://doi.org/10.1196/annals.1433.008
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2. https://doi.org/10.1186/1754-6834-3-2
Arevalo-Gallegos A, Ahmad Z, Asgher M, Parra-Saldivar R, Iqbal HMN (2017) Lignocellulose: a sustainable material to produce value-added products with a zero waste approach-a review. Int J Biol Macromol 99:308–318. https://doi.org/10.1016/j.ijbiomac.2017.02.097
Bhattarai K, Stalick WM, McKay S, Geme G, Bhattarai N (2011) Biofuel: an alternative to fossil fuel for alleviating world energy and economic crises. J Environ Sci Health A Tox Hazard Subst Environ Eng 46(12):1424–1442. https://doi.org/10.1080/10934529.2011.607042
Bowman MJ, Jordan DB, Vermillion KE, Braker JD, Moon J, Liu ZL (2010) Stereochemistry of furfural reduction by a Saccharomyces cerevisiae aldehyde reductase that contributes to in situ furfural detoxification. Appl Environ Microbiol 76(15):4926–4932. https://doi.org/10.1128/aem.00542-10
Brooks PJ, Theruvathu JA (2005) DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol 35(3):187–193. https://doi.org/10.1016/j.alcohol.2005.03.009
Cavka A, Stagge S, Jonsson LJ (2015) Identification of small aliphatic aldehydes in pretreated lignocellulosic feedstocks and evaluation of their inhibitory effects on yeast. J Agric Food Chem 63(44):9747–9754. https://doi.org/10.1021/acs.jafc.5b04803
Chang D, Yu Z (2018) Proteomic and metabolomic analysis of the cellular biomarkers related to inhibitors tolerance in Zymomonas mobilis ZM4. Biotechnol Biofuels 11:283. https://doi.org/10.1186/s13068-018-1287-5
Chen Y, Sheng J, Jiang T, Stevens J, Feng X, Wei N (2016) Transcriptional profiling reveals molecular basis and novel genetic targets for improved resistance to multiple fermentation inhibitors in Saccharomyces cerevisiae. Biotechnol Biofuels 9:9. https://doi.org/10.1186/s13068-015-0418-5
De Bhowmick G, Sarmah AK, Sen R (2018) Lignocellulosic biorefinery as a model for sustainable development of biofuels and value added products. Bioresour Technol 247:1144–1154. https://doi.org/10.1016/j.biortech.2017.09.163
Dellarco VL (1988) A mutagenicity assessment of acetaldehyde. Mutat Res 195(1):1–20. https://doi.org/10.1016/0165-1110(88)90013-9
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution -- principles and applications for biotechnology. Microb Cell Factories 12:64. https://doi.org/10.1186/1475-2859-12-64
Ehsani M, Fernandez MR, Biosca JA, Dequin S (2009a) Reversal of coenzyme specificity of 2,3-butanediol dehydrogenase from Saccharomyces cerevisae and in vivo functional analysis. Biotechnol Bioeng 104(2):381–389. https://doi.org/10.1002/bit.22391
Ehsani M, Fernandez MR, Biosca JA, Julien A, Dequin S (2009b) Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl Environ Microbiol 75(10):3196–3205. https://doi.org/10.1128/aem.02157-08
Gao J, Yang HH, Feng XH, Li S, Xu H (2013) A 2,3-butanediol dehydrogenase from Paenibacillus polymyxa ZJ-9 for mainly producing R,R-2,3-butanediol: purification, characterization and cloning. J Basic Microbiol 53(9):733–741. https://doi.org/10.1002/jobm.201200152
Gasch AP, Werner-Washburne M (2002) The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2(4–5):181–192. https://doi.org/10.1007/s10142-002-0058-2
Gonzalez E, Fernandez MR, Larroy C, Sola L, Pericas MA, Pares X, Biosca JA (2000) Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J Biol Chem 275(46):35876–35885. https://doi.org/10.1074/jbc.M003035200
Gonzalez E, Fernandez MR, Larroy C, Pares X, Biosca JA (2001) Characterization and functional role of Saccharomyces cerevisiae 2,3-butanediol dehydrogenase. Chem Biol Interact 130-132(1–3):425–434. https://doi.org/10.1016/s0009-2797(00)00282-9
Gonzalez E, Fernandez MR, Marco D, Calam E, Sumoy L, Pares X, Dequin S, Biosca JA (2010) Role of Saccharomyces cerevisiae oxidoreductases Bdh1p and Ara1p in the metabolism of acetoin and 2,3-butanediol. Appl Environ Microbiol 76(3):670–679. https://doi.org/10.1128/aem.01521-09
Ishida Y, Nguyen TT, Kitajima S, Izawa S (2016) Prioritized expression of BDH2 under bulk translational repression and its contribution to tolerance to severe vanillin stress in Saccharomyces cerevisiae. Front Microbiol 7:1059. https://doi.org/10.3389/fmicb.2016.01059
Ishida Y, Nguyen TTM, Izawa S (2017) The yeast ADH7 promoter enables gene expression under pronounced translation repression caused by the combined stress of vanillin, furfural, and 5-hydroxymethylfurfural. J Biotechnol 252:65–72. https://doi.org/10.1016/j.jbiotec.2017.04.024
Jacobs AT, Marnett LJ (2010) Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res 43(5):673–683. https://doi.org/10.1021/ar900286y
Jain M, Nagar P, Sharma A, Batth R, Aggarwal S, Kumari S, Mustafiz A (2018) GLYI and D-LDH play key role in methylglyoxal detoxification and abiotic stress tolerance. Sci Rep 8(1):5451. https://doi.org/10.1038/s41598-018-23806-4
Jayakody LN, Hayashi N, Kitagaki H (2011) Identification of glycolaldehyde as the key inhibitor of bioethanol fermentation by yeast and genome-wide analysis of its toxicity. Biotechnol Lett 33(2):285–292. https://doi.org/10.1007/s10529-010-0437-z
Jayakody LN, Ferdouse J, Hayashi N, Kitagaki H (2017) Identification and detoxification of glycolaldehyde, an unattended bioethanol fermentation inhibitor. Crit Rev Biotechnol 37(2):177–189. https://doi.org/10.3109/07388551.2015.1128877
Jonsson LJ, Alriksson B, Nilvebrant NO (2013) Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6(1):16. https://doi.org/10.1186/1754-6834-6-16
Karimi K, Taherzadeh MJ (2016) A critical review of analytical methods in pretreatment of lignocelluloses: composition, imaging, and crystallinity. Bioresour Technol 200:1008–1018. https://doi.org/10.1016/j.biortech.2015.11.022
Kim SJ, Seo SO, Jin YS, Seo JH (2013) Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresour Technol 146:274–281. https://doi.org/10.1016/j.biortech.2013.07.081
Ko JK, Um Y, Park YC, Seo JH, Kim KH (2015) Compounds inhibiting the bioconversion of hydrothermally pretreated lignocellulose. Appl Microbiol Biotechnol 99(10):4201–4212. https://doi.org/10.1007/s00253-015-6595-0
Kumar R, Singh S, Singh OV (2008) Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol 35(5):377–391. https://doi.org/10.1007/s10295-008-0327-8
Li BZ, Cheng JS, Ding MZ, Yuan YJ (2010) Transcriptome analysis of differential responses of diploid and haploid yeast to ethanol stress. J Biotechnol 148(4):194–203. https://doi.org/10.1016/j.jbiotec.2010.06.013
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90(3):809–825. https://doi.org/10.1007/s00253-011-3167-9
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81(4):743–753. https://doi.org/10.1007/s00253-008-1702-0
Moon J, Liu ZL (2015) Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae. Yeast 32(4):399–407. https://doi.org/10.1002/yea.3067
Noguchi C, Grothusen G, Anandarajan V, Martinez-Lage Garcia M, Terlecky D, Corzo K, Tanaka K, Nakagawa H, Noguchi E (2017) Genetic controls of DNA damage avoidance in response to acetaldehyde in fission yeast. Cell Cycle 16(1):45–58. https://doi.org/10.1080/15384101.2016.1237326
Nordling E, Jornvall H, Persson B (2002) Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur J Biochem 269(17):4267–4276. https://doi.org/10.1046/j.1432-1033.2002.03114.x
Pereira FB, Romani A, Ruiz HA, Teixeira JA, Domingues L (2014) Industrial robust yeast isolates with great potential for fermentation of lignocellulosic biomass. Bioresour Technol 161:192–199. https://doi.org/10.1016/j.biortech.2014.03.043
Persson B, Hedlund J, Jornvall H (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65(24):3879–3894. https://doi.org/10.1007/s00018-008-8587-z
Poudel S, Giannone RJ, Rodriguez M Jr, Raman B, Martin MZ, Engle NL, Mielenz JR, Nookaew I, Brown SD, Tschaplinski TJ, Ussery D, Hettich RL (2017) Integrated omics analyses reveal the details of metabolic adaptation of Clostridium thermocellum to lignocellulose-derived growth inhibitors released during the deconstruction of switchgrass. Biotechnol Biofuels 10:14. https://doi.org/10.1186/s13068-016-0697-5
Riveros-Rosas H, Julian-Sanchez A, Villalobos-Molina R, Pardo JP, Pina E (2003) Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur J Biochem 270(16):3309–3334. https://doi.org/10.1046/j.1432-1033.2003.03704.x
Sindhu R, Binod P, Pandey A (2016) Biological pretreatment of lignocellulosic biomass--an overview. Bioresour Technol 199:76–82. https://doi.org/10.1016/j.biortech.2015.08.030
Singh JK, Vyas P, Dubey A, Upadhyaya CP, Kothari R, Tyagi VV, Kumar A (2018) Assessment of different pretreatment technologies for efficient bioconversion of lignocellulose to ethanol. Front Biosci 10:350–371. https://doi.org/10.2741/s521
Skerker JM, Leon D, Price MN, Mar JS, Tarjan DR, Wetmore KM, Deutschbauer AM, Baumohl JK, Bauer S, Ibanez AB, Mitchell VD, Wu CH, Hu P, Hazen T, Arkin AP (2013) Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates. Mol Syst Biol 9:674. https://doi.org/10.1038/msb.2013.30
Sun S, Sun S, Cao X, Sun R (2016) The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour Technol 199:49–58. https://doi.org/10.1016/j.biortech.2015.08.061
Takusagawa Y, Otagiri M, Ui S, Ohtsuki T, Mimura A, Ohkuma M, Kudo T (2001) Purification and characterization of L-2,3-butanediol dehydrogenase of Brevibacterium saccharolyticum C-1012 expressed in Escherichia coli. Biosci Biotechnol Biochem 65(8):1876–1878. https://doi.org/10.1271/bbb.65.1876
Tan Y, Liu J, Liu Z, Li F (2014) Characterization of two novel butanol dehydrogenases involved in butanol degradation in syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528. J Basic Microbiol 54(9):996–1004. https://doi.org/10.1002/jobm.201300046
Tan Y, Liu ZY, Liu Z, Li FL (2015) Characterization of an acetoin reductase/2,3-butanediol dehydrogenase from Clostridium ljungdahlii DSM 13528. Enzym Microb Technol 79-80:1–7. https://doi.org/10.1016/j.enzmictec.2015.06.011
Thao MT, Gaillard ER (2016) The glycation of fibronectin by glycolaldehyde and methylglyoxal as a model for aging in Bruch’s membrane. Amino Acids 48(7):1631–1639. https://doi.org/10.1007/s00726-016-2222-3
Thompson HW, Mera R, Prasad C (1998) A description of the appropriate use of Student’s t-test. Nutr Neurosci 1(2):165–172. https://doi.org/10.1080/1028415X.1998.11747226
van der Pol EC, Bakker RR, Baets P, Eggink G (2014) By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio)chemicals and fuels. Appl Microbiol Biotechnol 98(23):9579–9593. https://doi.org/10.1007/s00253-014-6158-9
Vriesekoop F, Pamment NB (2005) Acetaldehyde addition and pre-adaptation to the stressor together virtually eliminate the ethanol-induced lag phase in Saccharomyces cerevisiae. Lett Appl Microbiol 41(5):424–427. https://doi.org/10.1111/j.1472-765X.2005.01777.x
Vriesekoop F, Barber AR, Pamment NB (2007) Acetaldehyde mediates growth stimulation of ethanol-stressed Saccharomyces cerevisiae: evidence of a redox-driven mechanism. Biotechnol Lett 29(7):1099–1103. https://doi.org/10.1007/s10529-007-9367-9
Wang X, Bai X, Chen DF, Chen FZ, Li BZ, Yuan YJ (2015) Increasing proline and myo-inositol improves tolerance of Saccharomyces cerevisiae to the mixture of multiple lignocellulose-derived inhibitors. Biotechnol Biofuels 8:142. https://doi.org/10.1186/s13068-015-0329-5
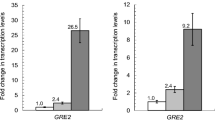
Wang X, Ma M, Liu ZL, Xiang Q, Li X, Liu N, Zhang X (2016) GRE2 from Scheffersomyces stipitis as an aldehyde reductase contributes tolerance to aldehyde inhibitors derived from lignocellulosic biomass. Appl Microbiol Biotechnol 100(15):6671–6682. https://doi.org/10.1007/s00253-016-7445-4
Wang HY, Xiao DF, Zhou C, Wang LL, Wu L, Lu YT, Xiang QJ, Zhao K, Li X, Ma M (2017) YLL056C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity. Appl Microbiol Biotechnol 101(11):4507–4520. https://doi.org/10.1007/s00253-017-8209-5
Wang H, Li Q, Kuang X, Xiao D, Han X, Hu X, Li X, Ma M (2018) Functions of aldehyde reductases from Saccharomyces cerevisiae in detoxification of aldehyde inhibitors and their biotechnological applications. Appl Microbiol Biotechnol 102(24):10439–10456. https://doi.org/10.1007/s00253-018-9425-3
Yi X, Gu H, Gao Q, Liu ZL, Bao J (2015) Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels 8:153. https://doi.org/10.1186/s13068-015-0333-9
Yu M, Huang M, Song Q, Shao J, Ying X (2015) Characterization of a (2R,3R)-2,3-butanediol dehydrogenase from Rhodococcus erythropolis WZ010. Molecules 20(4):7156–7173. https://doi.org/10.3390/molecules20047156
Zhang K, Tong M, Gao K, Di Y, Wang P, Zhang C, Wu X, Zheng D (2015) Genomic reconstruction to improve bioethanol and ergosterol production of industrial yeast Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 42(2):207–218. https://doi.org/10.1007/s10295-014-1556-7
Zhao X, Tang J, Wang X, Yang R, Zhang X, Gu Y, Li X, Ma M (2015) YNL134C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity for detoxification of furfural derived from lignocellulosic biomass. Yeast 32(5):409–422. https://doi.org/10.1002/yea.3068
Zhou Q, Liu ZL, Ning K, Wang A, Zeng X, Xu J (2014) Genomic and transcriptome analyses reveal that MAPK- and phosphatidylinositol-signaling pathways mediate tolerance to 5-hydroxymethyl-2-furaldehyde for industrial yeast Saccharomyces cerevisiae. Sci Rep 4:6556. https://doi.org/10.1038/srep06556
Acknowledgments
The authors thank James Allen from Liwen Bianji, Edanz Group China for revising this manuscript.
Funding
This work was funded by the Science and Technology Department of Sichuan Province (No. 2020YFH0142), the National Natural Science Foundation of China (No. 31570086), and the Talent Introduction Fund of Sichuan Agricultural University (No. 01426100).
Author information
Authors and Affiliations
Contributions
MGM, XLK, and YDOY conceived and designed research. XLK, YDOY, YPG, QL, and HYW conducted experiments. XLK, YDOY, and YPG analyzed data. YFG, XL, and QC contributed analytical tools. XLK and YDOY wrote the manuscript. MGM, GTA, and EA revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 447 kb).
Rights and permissions
About this article
Cite this article
Kuang, X., Ouyang, Y., Guo, Y. et al. New insights into two yeast BDHs from the PDH subfamily as aldehyde reductases in context of detoxification of lignocellulosic aldehyde inhibitors. Appl Microbiol Biotechnol 104, 6679–6692 (2020). https://doi.org/10.1007/s00253-020-10722-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10722-9