Abstract
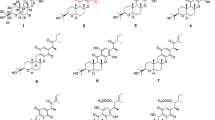
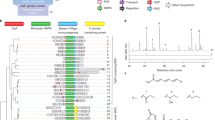
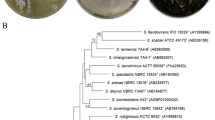
Halogen substituents are important for biological activity in many compounds. Genome-based mining of halogenase along with its biosynthetic gene cluster provided an efficient approach for the discovery of naturally occurring organohalogen compounds. Analysis of the genome sequence of a phytopathogenic fungus Bipolaris sorokiniana 11134 revealed a polyketide gene cluster adjacent to a flavin-dependent halogenase capable of encoding halogenated polyketides, which are rarely reported in phytopathogenic fungi. Furthermore, MS- and UV-guided isolation and purification led to the identification of five chlorine-containing natural products together with seven other chromones and xanthones. Two of the chlorinated compounds and four chromones are new compounds. Their structures were elucidated by NMR spectroscopic analysis and HRESIMS data. The biosynthetic gene clusters of isolated compounds and their putative biosynthetic pathway are also proposed. One new chlorinated compound showed activity against Staphylococcus aureus, methicillin-resistant S. aureus, and three clinical-resistant S. aureus strains with a shared minimum inhibitory concentration (MIC) of 12.5 μg/mL. Genome-based mining of halogenases combined with high-resolution MS- and UV-guided identification provides an efficient approach to discover new halogenated natural products from microorganisms.





Similar content being viewed by others
References
Abdel-Mageed WM, Bayoumi SA, Al-wahaibi LH, Li L, Sayed HM, Abdelkader MS, El-Gamal AA, Liu M, Zhang J, Zhang L, Liu X (2016) Noncyanogenic cyanoglucoside cyclooxygenase inhibitors from Simmondsia chinensis. Org Lett 18(8):1728–1731. https://doi.org/10.1021/acs.orglett.6b00206
Abdelmohsen UR, Balasubramanian S, Oelschlaeger TA, Grkovic T, Pham NB, Quinn RJ, Hentschel U (2017) Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect Dis 17(2):e30–e41. https://doi.org/10.1016/S1473-3099(16)30323-1
Agarwal V, Miles ZD, Winter JM, Eustáquio AS, El Gamal AA, Moore BS (2017) Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem Rev 117(8):5619–5674. https://doi.org/10.1021/acs.chemrev.6b00571
Aich S, Singh RK, Kundu P, Pandey SP, Datta S (2017) Genome-wide characterization of cellulases from the hemi-biotrophic plant pathogen, Bipolaris sorokiniana, reveals the presence of a highly stable GH7 endoglucanase. Biotechnol Biofuels 10(1):135. https://doi.org/10.1186/s13068-017-0822-0
Ansari MZ, Yadav G, Gokhale RS, Mohanty D (2004) NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res 32(suppl_2):W405–W413. https://doi.org/10.1093/nar/gkh359
Bachmann BO, Ravel J (2009) Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol 458:181–217. https://doi.org/10.1016/S0076-6879(09)04808-3
Bao J, Luo J-F, Qin X-C, Xu X-Y, Zhang X-Y, Tu Z-C, Qi S-H (2014) Dihydrothiophene-condensed chromones from a marine-derived fungus Penicillium oxalicum and their structure–bioactivity relationship. Bioorg Med Chem Lett 24(11):2433–2436. https://doi.org/10.1016/j.bmcl.2014.04.028
Birney E, Clamp M, Durbin R (2004) GeneWise and genomewise. Genome Res 14(5):988–995. https://doi.org/10.1101/gr.1865504
Bohnert M, Nützmann H-W, Schroeckh V, Horn F, Dahse H-M, Brakhage AA, Hoffmeister D (2014) Cytotoxic and antifungal activities of melleolide antibiotics follow dissimilar structure–activity relationships. Phytochemistry 105:101–108. https://doi.org/10.1016/j.phytochem.2014.05.009
Boufridi A, Quinn RJ (2018) Harnessing the properties of natural products. Annu Rev Pharmacol Toxicol 58:451–470. https://doi.org/10.1146/annurev-pharmtox-010716-105029
Bringmann G, Irmer A, Feineis D, Gulder TA, Fiedler H-P (2009) Convergence in the biosynthesis of acetogenic natural products from plants, fungi, and bacteria. Phytochemistry 70(15–16):1776–1786. https://doi.org/10.1016/j.phytochem.2009.08.019
Carlson H, Nilsson P, Jansson H, Odham G (1991) Characterization and determination of prehelminthosporol, a toxin from the plant pathogenic fungus Bipolaris sorokiniana, using liquid chromatography/mass spectrometry. J Microbiol Methods 13(4):259–269. https://doi.org/10.1016/0167-7012(91)90063-V
Chiang Y-M, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CC, Oakley BR (2010) Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol 76(7):2067–2074. https://doi.org/10.1128/AEM.02187-09
Chooi Y-H, Cacho R, Tang Y (2010) Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol 17(5):483–494. https://doi.org/10.1016/j.chembiol.2010.03.015
Condon BJ, Leng Y, Wu D, Bushley KE, Ohm RA, Otillar R, Martin J, Schackwitz W, Grimwood J, MohdZainudin N (2013) Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet 9(1):e1003233. https://doi.org/10.1371/journal.pgen.1003233
Dashti Y, Grkovic T, Abdelmohsen UR, Hentschel U, Quinn RJ (2017) Actinomycete metabolome induction/suppression with N-acetylglucosamine. J Nat Prod 80(4):828–836. https://doi.org/10.1021/acs.jnatprod.6b00673
Demain AL, Zhang L (2005) Natural products and drug discovery. In: Natural products. Springer, Berlin, pp 3–29
Ferrara M, Perrone G, Gambacorta L, Epifani F, Solfrizzo M, Gallo A (2016) Identification of a halogenase involved in the biosynthesis of ochratoxin A in Aspergillus carbonarius. Appl Environ Microbiol 82(18):5631–5641. https://doi.org/10.1128/AEM.01209-16
Flor H (1955) Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology 45:680–685
Gaudêncio SP, Pereira F (2015) Dereplication: racing to speed up the natural products discovery process. Nat Prod Rep 32(6):779–810. https://doi.org/10.1039/c4np00134f
Gribble GW (2004) Natural organohalogens: a new frontier for medicinal agents? J Chem Educ 81(10):1441. https://doi.org/10.1021/ed081p1441
Gribble GW (2015) A recent survey of naturally occurring organohalogen compounds. Environ Chem 12(4):396–405. https://doi.org/10.1071/EN15002
Gribble GW, Scheuer PJ, Moore RE, Faulkner DJ (2018) Newly discovered naturally occurring organohalogens. Arkivoc(VI):0–0. https://doi.org/10.24820/ark.5550190.p010.610
Haridas S, Breuill C, Bohlmann J, Hsiang T (2011) A biologist’s guide to de novo genome assembly using next-generation sequence data: a test with fungal genomes. J Microbiol Methods 86(3):368–375. https://doi.org/10.1016/j.mimet.2011.06.019
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14(2):111–129. https://doi.org/10.1038/nrd4510
He W, Liu M, Huang P, Abdel-Mageed WM, Han J, Watrous JD, Nguyen DD, Wang W, Song F, Dai H, Zhang J, Quinn RJ, Grkovic T, Luo H, Zhang L, Liu X (2016a) Discovery of tanshinone derivatives with anti-MRSA activity via targeted bio-transformation. Synth Syst Biotechnol 1(3):187–194. https://doi.org/10.1016/j.synbio.2016.05.002
He W, Liu M, Li X, Zhang X, Abdel-Mageed WM, Li L, Wang W, Zhang J, Han J, Dai H, Quinn RJ, H-w L, Luo H, Zhang L, Liu X (2016b) Fungal biotransformation of tanshinone results in [4 + 2] cycloaddition with sorbicillinol: evidence for enzyme catalysis and increased antibacterial activity. Appl Microbiol Biotechnol 100(19):8349–8357. https://doi.org/10.1007/s00253-016-7488-6
He W, Li Y, Qin Y, Tong X, Song Z, Zhao Y, Wei R, Li L, Dai H, Wang W, Luo H, Ye X, Zhang L, Liu X (2017) New cryptotanshinone derivatives with anti-influenza A virus activities obtained via biotransformation by Mucor rouxii. Appl Microbiol Biotechnol 101(16):6365–6374. https://doi.org/10.1007/s00253-017-8351-0
Hiramatsu K, Cui L, Kuroda M, Ito T (2001) The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9(10):486–493. https://doi.org/10.1016/S0966-842X(01)02175-8
Horiguchi K, Suzuki Y, Sassa T (1989) Biosynthetic study of chloromonilicin, a growth self-inhibitor having a novel lactone ring, from Monilinia fructicola. Agric Biol Chem 53(8):2141–2145. https://doi.org/10.1271/bbb1961.53.2141
Huang P, Xie F, Ren B, Wang Q, Wang J, Wang Q, Abdel-Mageed WM, Liu M, Han J, Oyeleye A, Shen J, Song F, Dai HQ, Liu X, Zhang L (2016) Anti-MRSA and anti-TB metabolites from marine-derived Verrucosispora sp. MS100047. Appl Microbiol Biotechnol 100(17):7437–7447. https://doi.org/10.1007/s00253-016-7406-y
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36(suppl_2):W5–W9. https://doi.org/10.1093/nar/gkn201
Kachi H, Sassa T (1986) Isolation of moniliphenone, a key intermediate in xanthone biosynthesis from Monilinia fructicola. Agric Biol Chem 50(6):1669–1671. https://doi.org/10.1080/00021369.1986.10867632
Kachi H, Attori H, Assa T (1986) A new antifungal substance, bromomonilicin, and its precursor produced by Monilinia fructicola. J Antibiot 39(1):164–166. https://doi.org/10.7164/antibiotics.39.164
Karaiskos I, Giamarellou H (2014) Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 15(10):1351–1370. https://doi.org/10.1517/14656566.2014.914172
Kenfield D, Bunkers G, Strobel GA, Sugawara F (1988) Potential new herbicides–phytotoxins from plant pathogens. Weed Technol 2(4):519–524. https://doi.org/10.1017/S0890037X00032383
Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298(15):1763–1771. https://doi.org/10.1001/jama.298.15.1763
Kurtböke Dİ, French JR, Hayes RA, Quinn RJ (2014) Eco-taxonomic insights into actinomycete symbionts of termites for discovery of novel bioactive compounds. In: Biotechnological applications of biodiversity. Springer, Berlin, pp 111–135
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Law K-K, Chan T-L, Tam SW, Shatin N (1979) Synthesis of pinselic acid and pinselin. J Org Chem 44(24):4452–4453
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24(5):713–714. https://doi.org/10.1093/bioinformatics/btn025
Liu M, Grkovic T, Liu X, Han J, Zhang L, Quinn RJ (2017a) A systems approach using OSMAC, log P and NMR fingerprinting: an approach to novelty. Synthetic and Systems Biotechnology 2(4):276–286. https://doi.org/10.1016/j.synbio.2017.10.001
Liu M, Huang P, Wang Q, Ren B, Oyeleye A, Liu M, Zhang J, Li X, Zhang X, Zhang L, Liu X (2017b) Synergistic antifungal indolecarbazoles from Streptomyces sp. CNS-42 associated with traditional Chinese medicine Alisma orientale. J Antibiot 70(5):715–717. https://doi.org/10.1038/ja.2016.160
Liu X, Xie F, Doughty LB, Wang Q, Zhang L, Liu X, Cheng Y-Q (2018) Genomics-guided discovery of a new and significantly better source of anticancer natural drug FK228. Synth Syst Biotechnol 3(4):268–274. https://doi.org/10.1016/j.synbio.2018.10.011
Ma TT, Shan WG, Ying YM, Ma LF, Liu WH, Zhan ZJ (2015) Xanthones with α-glucosidase inhibitory activities from Aspergillus versicolor, a fungal endophyte of Huperzia serrata. Helv Chim Acta 98(1):148–152. https://doi.org/10.1002/hlca.201400165
Maes CM, Steyn PS, Vleggaar R, Kirby GW, Robins DJ, Stark WM (1985) Structure and biosynthesis of bipolaramide, a novel dioxopiperazine from Bipolaris sorokiniana. J Chem Soc, Perkin Trans 1:2489–2492. https://doi.org/10.1039/P19850002489
Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, Harbarth S, Hindler J, Kahlmeter G, Olsson-Liljequist B (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Malaker P, Mian L, Khandaker MM, Reza M (2007) Survival of Bipolaris sorokiniana (Sacc.) Shoemaker in soil and residue of wheat. Bangladesh J Botany 36(2):133–137. https://doi.org/10.3329/bjb.v36i2.1501
Masi M, Meyer S, Clement S, Pescitelli G, Cimmino A, Cristofaro M, Evidente A (2017) Chloromonilinic acids C and D, phytotoxic tetrasubstituted 3-chromanonacrylic acids isolated from Cochliobolus australiensis with potential herbicidal activity against buffelgrass (Cenchrus ciliaris). J Nat Prod 80(10):2771–2777. https://doi.org/10.1021/acs.jnatprod.7b00583
McDonald MC, Ahren D, Simpfendorfer S, Milgate A, Solomon PS (2018) The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol Plant Pathol 19(2):432–439. https://doi.org/10.1111/mpp.12535
Minerdi D, Zgrablic I, Castrignanò S, Catucci G, Medana C, Terlizzi ME, Gribaudo G, Gilardi G, Sadeghi SJ (2016) Escherichia coli overexpressing a Baeyer-Villiger monooxygenase from Acinetobacter radioresistens becomes resistant to imipenem. Antimicrob Agents Chemother 60(1):64–74. https://doi.org/10.1128/AAC.01088-15
Miyagawa H, Nagai S, Tsurushima T, Sato M, Ueno T, Fukami H (1994) Phytotoxins produced by the plant pathogenic fungus Bipolaris bicolor El-1. Biosci Biotechnol Biochem 58(6):1143–1145. https://doi.org/10.1271/bbb.58.1143
Murphy CD (2006) Recent developments in enzymatic chlorination. Nat Prod Rep 23(2):147–152. https://doi.org/10.1039/b516588c
Neumann CS, Fujimori DG, Walsh CT (2008) Halogenation strategies in natural product biosynthesis. Chem Biol 15(2):99–109. https://doi.org/10.1016/j.chembiol.2008.01.006
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79(3):629–661. https://doi.org/10.1021/acs.jnatprod.5b01055
Phuwapraisirisan P, Sawang K, Siripong P, Tip-Pyang S (2007) Anhydrocochlioquinone a, a new antitumor compound from Bipolaris oryzae. Tetrahedron Lett 48(30):5193–5195. https://doi.org/10.1016/j.tetlet.2007.05.151
Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG (2017) Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci U S A 114(22):5601–5606. https://doi.org/10.1073/pnas.1614680114
Qi QY, Huang L, He LW, Han JJ, Chen Q, Cai L, Liu HW (2014) Cochlioquinone derivatives with apoptosis-inducing effects on HCT116 colon cancer cells from the phytopathogenic fungus Bipolaris luttrellii L439. Chem Biodivers 11(12):1892–1899. https://doi.org/10.1002/cbdv.201400106
Quinn RJ, Janso JE (2011) Recent developments in natural products: potential impact on antibacterial drug discovery. Emerging trends in antibacterial discovery: answering the call to arms, ed by AA Miller, PF Miller (Caister Academic, Norfolk 2011) pp:149–169
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425 https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sassa T, Kachi H, Nukina M, Suzuki Y (1985) Chloromonilicin, a new antifungal metabolite produced by Monilinia fructicola. J Antibiot 38(3):439–441. https://doi.org/10.7164/antibiotics.38.439
Sassa T, Horiguchi K, Suzuki Y (1989) Chloromonilinic acids A and B, novel catabolites of the growth self-inhibitor chloromonilicin isolated from Monilinia fructicola. Agric Biol Chem 53(5):1337–1341. https://doi.org/10.1080/00021369.1989.10869453
Schwab JM, Li W, Thomas LP (1983) Cyclohexanone oxygenase: stereochemistry, enantioselectivity, and regioselectivity of an enzyme-catalyzed Baeyer-Villiger reaction. J Am Chem Soc 105(14):4800–4808. https://doi.org/10.1021/ja00352a044
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19(6):1117–1123. https://doi.org/10.1101/gr.089532.108
Sivanesan A (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. CAB International, Wallingford
Song F, Ren B, Yu K, Chen C, Guo H, Yang N, Gao H, Liu X, Liu M, Tong Y, Dai HQ, Bai H, Wang J, Zhang L (2012) Quinazolin-4-one coupled with pyrrolidin-2-iminium alkaloids from marine-derived fungus Penicillium aurantiogriseum. Mar Drugs 10(6):1297–1306. https://doi.org/10.3390/md10061297
Sugawara F, Strobel G, Fisher L, Van Duyne G, Clardy J (1985) Bipolaroxin, a selective phytotoxin produced by Bipolaris cynodontis. Proc Natl Acad Sci U S A 82(24):8291–8294. https://doi.org/10.1073/pnas.82.24.8291
Sun Y-L, He F, Liu K-S, Zhang X-Y, Bao J, Wang Y-F, Nong X-H, Xu X-Y, Qi S-H (2012) Cytotoxic dihydrothiophene-condensed chromones from marine-derived fungus Penicillium oxalicum. Planta Med 78(18):1957–1961. https://doi.org/10.1055/s-0032-1327874
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Taneja NK, Tyagi JS (2007) Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J Antimicrob Chemother 60(2):288–293. https://doi.org/10.1093/jac/dkm207
van Pée K-H, Patallo EP (2006) Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl Microbiol Biotechnol 70(6):631–641. https://doi.org/10.1007/s00253-005-0232-2
Wang S, Xu Y, Maine EA, Wijeratne EK, Espinosa-Artiles P, Gunatilaka AL, Molnár I (2008) Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol 15(12):1328–1338. https://doi.org/10.1016/j.chembiol.2008.10.006
Wang JF, Dai HQ, Wei YL, Zhu HJ, Yan YM, Wang YH, Long CL, Zhong HM, Zhang LX, Cheng YX (2010) Antituberculosis agents and an inhibitor of the para-aminobenzoic acid biosynthetic pathway from hydnocarpus anthelminthica seeds. Chem Biodivers 7(8):2046–2053. https://doi.org/10.1002/cbdv.201000072
Wang Q, Song F, Xiao X, Huang P, Li L, Monte A, Abdel-Mageed WM, Wang J, Guo H, He W, Xie F, Dai HQ, Liu M, Chen C, Xu H, Liu M, Piggott AM, Liu X, Capon RJ, Zhang L (2013) Abyssomicins from the South China Sea deep-sea sediment Verrucosispora sp.: natural Thioether Michael addition adducts as antitubercular prodrugs. Angew Chem Int Ed 52(4):1231–1234. https://doi.org/10.1002/anie.201208801
Wang F, Wang Y, Ji J, Zhou Z, Yu J, Zhu H, Su Z, Zhang L, Zheng J (2015) Structural and functional analysis of the loading acyltransferase from avermectin modular polyketide synthase. ACS Chem Biol 10(4):1017–1025. https://doi.org/10.1021/cb500873k
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43(W1):W237–W243. https://doi.org/10.1093/nar/gkv437
Wick J, Heine D, Lackner G, Misiek M, Tauber J, Jagusch H, Hertweck C, Hoffmeister D (2016) A fivefold parallelized biosynthetic process secures chlorination of Armillaria mellea (honey mushroom) toxins. Appl Environ Microbiol 82(4):1196–1204. https://doi.org/10.1128/AEM.03168-15
Wright GD (2017) Opportunities for natural products in 21 st century antibiotic discovery. Nat Prod Rep 34(7):694–701. https://doi.org/10.1039/C7NP00019G
Xu X, Liu L, Zhang F, Wang W, Li J, Guo L, Che Y, Liu G (2014) Identification of the first diphenyl ether gene cluster for pestheic acid biosynthesis in plant endophyte Pestalotiopsis fici. ChemBioChem 15(2):284–292. https://doi.org/10.1002/cbic.201300626
Yadav G, Gokhale RS, Mohanty D (2003) SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res 31(13):3654–3658. https://doi.org/10.1093/nar/gkg607
Yadav G, Gokhale RS, Mohanty D (2009) Towards prediction of metabolic products of polyketide synthases: an in silico analysis. PLoS Comp Biol 5(4):e1000351. https://doi.org/10.1371/journal.pcbi.1000351
Yan W, Song H, Song F, Guo Y, Wu C-H, Her AS, Pu Y, Wang S, Naowarojna N, Weitz A, HM P, CC E, Zhang L, Liu P, Zhang YJ (2015) Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme. Nature 527(7579):539–543. https://doi.org/10.1038/nature15519
Zeng J, Zhan J (2010) A novel fungal flavin-dependent halogenase for natural product biosynthesis. ChemBioChem 11(15):2119–2123. https://doi.org/10.1002/cbic.201000439
Zerbino D, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Zhang L, An R, Wang J, Sun N, Zhang S, Hu J, Kuai J (2005) Exploring novel bioactive compounds from marine microbes. Curr Opin Microbiol 8(3):276–281. https://doi.org/10.1016/j.mib.2005.04.008
Zhang J, Abdel-Mageed WM, Liu M, Huang P, He W, Li L, Song F, Dai H, Liu X, Liang J, Zhang L (2013) Caesanines A–D, new cassane diterpenes with unprecedented N bridge from Caesalpinia sappan. Org Lett 15(18):4726–4729. https://doi.org/10.1021/ol402058z
Funding
This work was supported by grants from the National Natural Science Foundation of China (31430002, 81573341, 21877038, 31720103901, 31320103911), Open Project Funding of the State Key Laboratory of Bioreactor Engineering, the 111 Project (B18022), the Fundamental Research Funds for the Central Universities (22221818014), the Natural Science Foundation from Shandong Province (No. ZR2017ZB0206), and Taishan Scholarship. The authors acknowledge the Australian Research Council (ARC) for support (LE140100119, LE20100170). J. Han thanks the Griffith University for the provision of the Ph.D. scholarships (GUPRS and GUIPRS). Genome sequencing and assembly of BS11134 were supported by funding from the Natural Science and Engineering Research Council of Canada to T. Hsiang.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 2207 kb)
Rights and permissions
About this article
Cite this article
Han, J., Zhang, J., Song, Z. et al. Genome- and MS-based mining of antibacterial chlorinated chromones and xanthones from the phytopathogenic fungus Bipolaris sorokiniana strain 11134. Appl Microbiol Biotechnol 103, 5167–5181 (2019). https://doi.org/10.1007/s00253-019-09821-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09821-z