Abstract
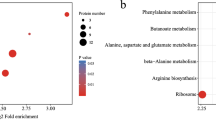
The pseudonocardiate Amycolatopsis sp. ATCC 39116 is used for the biotechnical production of natural vanillin from ferulic acid. Our laboratory has performed genetic modifications of this strain previously, but there are still many gaps in our knowledge regarding its vanillin tolerance and the general metabolism. We performed cultivations with this bacterium and compared the proteomes of stationary phase cells before ferulic acid feeding with those during ferulic acid feeding. Thereby, we identified 143 differently expressed proteins. Deletion mutants were constructed and characterized to analyze the function of nine corresponding genes. Using these mutants, we identified an active ferulic acid β-oxidation pathway and the enzymes which constitute this pathway. A combined deletion mutant in which the β-oxidation as well as non-β-oxidation pathways of ferulic acid degradation were deleted was unable to grow on ferulic acid as the sole source of carbon and energy. This mutant differs from the single deletion mutants and was unable to grow on ferulic acid. Furthermore, we showed that the non-β-oxidation pathway is involved in caffeic acid degradation; however, its deletion is complemented even in the double deletion mutant. This shows that both pathways can complement each other. The β-oxidation deletion mutant produced significantly reduced amounts of vanillic acid (0.12 instead of 0.35 g/l). Therefore, the resulting mutant could be used as an improved production strain. The quinone oxidoreductase deletion mutant (ΔytfG) degraded ferulic acid slower at first but produced comparable amounts of vanillin and significantly less vanillyl alcohol when compared to the parent strain.








Similar content being viewed by others
References
Achterholt S, Priefert H, Steinbüchel A (2000) Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl Microbiol Biotechnol 54:799–807. https://doi.org/10.1007/s002530000431
Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister JR, Bryant SH, Canese K, Charowhas C, Clark K, Dicuccio M, Dondoshansky I, Federhen S, Feolo M, Funk K, Geer LY, Gorelenkov V, Hoeppner M, Holmes B, Johnson M, Khotomlianski V, Kimchi A, Kimelman M, Kitts P, Klimke W, Krasnov S, Kuznetsov A, Landrum MJ, Landsman D, Lee JM, Lipman DJ, Lu Z, Madden TL, Madej T, Marchler-Bauer A, Karsch-Mizrachi I, Murphy T, Orris R, Ostell J, O’sullivan C, Panchenko A, Phan L, Preuss D, Pruitt KD, Rodarmer K, Rubinstein W, Sayers E, Schneider V, Schuler GD, Sherry ST, Sirotkin K, Siyan K, Slotta D, Soboleva A, Soussov V, Starchenko G, Tatusova TA, Todorov K, Trawick BW, Vakatov D, Wang Y, Ward M, Wilbur WJ, Yaschenko E, Zbicz K (2016) Database resources of the National Center for Biotechnology Information. Nucl Acids Res 44:D7–D19. https://doi.org/10.1093/nar/gkv1290
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Alves AMCR, Euverink GJW, Hektor HJ, Hessels GI, van der Vlag J, Vrijbloed JW, Hondmann D, Visser J, Dijkhuizen L (1994) Enzymes of glucose and methanol metabolism in the actinomycete Amycolatopsis methanolica. J Bacteriol 176:6827–6835. https://doi.org/10.1128/jb.176.22.6827-6835.1994
Alves AMCR, Euverink GJW, Bibb MJ, Dijkhuizen L (1997) Identification of ATP-dependent phosphofructokinase as a regulatory step in the glycolytic pathway of the actinomycete Streptomyces coelicolor A3(2). Appl Environ Microbiol 63:956–961
Alves AMCR, Euverink GJW, Santos H, Dijkhuizen L (2001) Different physiological roles of ATP- and PPi-dependent phosphofructokinase isoenzymes in the methylotrophic actinomycete Amycolatopsis methanolica. J Bacteriol 183:7231–7240. https://doi.org/10.1128/JB.183.24.7231-7240.2001
Andreoni V, Bestetti G (1986) Comparative analysis of different Pseudomonas strains that degrade cinnamic acid. Appl Environ Microbiol 52:930–934
Barnes MR, Duetz WA, Williams PA (1997) A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: cloning and characterization of the hpp operon. J Bacteriol 179:6145–6153
Bennett JP, Bertin L, Moulton B, Fairlamb IJS, Brzozowski AM, Walton NJ, Grogan G (2008) A ternary complex of hydroxycinnamoyl-CoA hydratase–lyase (HCHL) with acetyl-CoA and vanillin gives insights into substrate specificity and mechanism. Biochem J 414:281–289. https://doi.org/10.1042/BJ20080714
Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300
Beste DJV, Bonde B, Hawkins N, Ward JL, Beale MH, Noack S, Nöh K, Kruger NJ, Ratcliffe RG, McFadden J (2011) 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in Mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog 7:e1002091. https://doi.org/10.1371/journal.ppat.1002091
Borodina I, Schöller C, Eliasson A, Nielsen J (2005) Metabolic network analysis of Streptomyces tenebrarius, a Streptomyces species with an active Entner-Doudoroff pathway. Appl Environ Microbiol 71:2294–2302. https://doi.org/10.1128/AEM.71.5.2294-2302.2005
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Brandt U, Waletzko C, Voigt B, Hecker M, Steinbüchel A (2014) Mercaptosuccinate metabolism in Variovorax paradoxus strain B4—a proteomic approach. Appl Microbiol Biotechnol 98:6039–6050. https://doi.org/10.1007/s00253-014-5811-7
Brochado AR, Matos C, Møller BL, Hansen J, Mortensen UH, Patil KR (2010) Improved vanillin production in baker’s yeast through in silico design. Microb Cell Factories 9:84. https://doi.org/10.1186/1475-2859-9-84
Campillo T, Renoud S, Kerzaon I, Vial L, Baude J, Gaillard V, Bellvert F, Chamignon C, Comte G, Nesme X, Lavire C, Hommais F (2014) Analysis of hydroxycinnamic acid degradation in Agrobacterium fabrum reveals a coenzyme A-dependent, beta-oxidative deacetylation pathway. Appl Environ Microbiol 80:3341–3349. https://doi.org/10.1128/AEM.00475-14
Chen H-P, Chow M, Liu C-C, Lau A, Liu J, Eltis LD (2012) Vanillin catabolism in Rhodococcus jostii RHA1. Appl Environ Microbiol 78:586–588. https://doi.org/10.1128/AEM.06876-11
Chen C, Pan J, Yang X, Guo C, Ding W, Si M, Zhang Y, Shen X, Wang Y (2016) Global transcriptomic analysis of the response of Corynebacterium glutamicum to vanillin. PLoS One 11:1–16. https://doi.org/10.1371/journal.pone.0164955
Chen C, Pan J, Yang X, Xiao H, Zhang Y, Si M, Shen X, Wang Y (2017) Global transcriptomic analysis of the response of Corynebacterium glutamicum to ferulic acid. Arch Microbiol 199:325–334. https://doi.org/10.1007/s00203-016-1306-5
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. https://doi.org/10.1126/science.1175371
Costa DJS, Kothe E, Abate CM, Amoroso MJ (2012) Unraveling the Amycolatopsis tucumanensis copper-resistome. Biometals 25:905–917. https://doi.org/10.1007/s10534-012-9557-3
Crosas E, Porté S, Moeini A, Farrés J, Biosca JA, Parés X, Fernández MR (2011) Novel alkenal/one reductase activity of yeast NADPH:quinone reductase Zta1p. Prospect of the functional role for the ζ-crystallin family. Chem Biol Interact 191:32–37. https://doi.org/10.1016/j.cbi.2011.01.021
Davis JR, Goodwin LA, Woyke T, Teshima H, Bruce D, Detter C, Tapia R, Han S, Han J, Pitluck S, Nolan M, Mikhailova N, Land ML, Sello JK (2012) Genome sequence of Amycolatopsis sp. strain ATCC 39116, a plant biomass-degrading actinomycete. J Bacteriol 194:2396–2397. https://doi.org/10.1128/JB.00186-12
Ehira S, Ogino H, Teramoto H, Inui M, Yukawa H (2009) Regulation of quinone oxidoreductase by redox-sensing transcriptional regulator QorR in Corynebacterium glutamicum. J Biol Chem 284:16736–16742. https://doi.org/10.1074/jbc.M109.009027
Falconnier B, Lapierre C, Lesage-Meessen L, Yonnet G, Brunerie P, Colonna-Ceccaldi B, Corrieu G, Asther M (1994) Vanillin as a product of ferulic acid biotransformation by the white-rot fungus Pycnoporus cinnabarinus I-937: identification of metabolic pathways. J Biotechnol 37:123–132. https://doi.org/10.1016/0168-1656(94)90003-5
Fleige C, Hansen G, Kroll J, Steinbüchel A (2013) Investigation of the Amycolatopsis sp. strain ATCC 39116 vanillin dehydrogenase and its impact on the biotechnical production of vanillin. Appl Environ Microbiol 79:81–90. https://doi.org/10.1128/AEM.02358-12
Fleige C, Steinbüchel A (2014) Construction of expression vectors for metabolic engineering of the vanillin-producing actinomycete Amycolatopsis sp. ATCC 39116. Appl Microbiol Biotechnol 98:6387–6395. https://doi.org/10.1007/s00253-014-5724-5
Fleige C, Meyer F, Steinbüchel A (2016) Metabolic engineering of the actinomycete Amycolatopsis sp. strain ATCC 39116 towards enhanced production of natural vanillin. Appl Environ Microbiol 82:3410–3419. https://doi.org/10.1128/AEM.00802-16
Foldes M, Munro R, Sorrell TC, Shanker S, Toohey M (1983) In-vitro effects of vancomycin, rifampicin, and fusidic acid, alone and in combination, against methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 11:21–26. https://doi.org/10.1093/jac/11.1.21
Frey UH, Bachmann HS, Peters J, Siffert W (2008) PCR-amplification of GC-rich regions: “slowdown PCR”. Nat Protoc 3:1312–1317. https://doi.org/10.1038/nprot.2008.112
Funahashi A, Tanimura N, Morohashi M, Kitano H (2003) CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico 1:159–162. https://doi.org/10.1016/S1478-5382(03)02370-9
Gallo G, Renzone G, Alduina R, Stegmann E, Weber T, Lantz AE, Thykaer J, Sangiorgi F, Scaloni A, Puglia AM (2010) Differential proteomic analysis reveals novel links between primary metabolism and antibiotic production in Amycolatopsis balhimycina. Proteomics 10:1336–1358. https://doi.org/10.1186/1475-2859-9-95
Hanahan D, Jessee J, Bloom RF (1991) Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204:63–113. https://doi.org/10.1016/0076-6879(91)04006-A
Hansen EH, Møller BL, Kock GR, Bünner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS, Hansen J (2009) De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl Environ Microbiol 75:2765–2774. https://doi.org/10.1128/AEM.02681-08
Hiller K, Grote A, Maneck M, Münch R, Jahn D (2006) JVirGel 2.0: computational prediction of proteomes separated via two-dimensional gel electrophoresis under consideration of membrane and secreted proteins. Bioinformatics 22:2441–2443. https://doi.org/10.1093/bioinformatics/btl409
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, Mcanulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJA, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C (2009) InterPro: the integrative protein signature database. Nucl Acids Res 37:211–215. https://doi.org/10.1093/nar/gkn785
Joseph P, Long DJ, Klein-Szanto AJ, Jaiswal AK (2000) Role of NAD(P)H:quinone oxidoreductase 1 (DT diaphorase) in protection against quinone toxicity. Biochem Pharmacol 60:207–214. https://doi.org/10.1016/S0006-2952(00)00321-X
Kallscheuer N, Vogt M, Kappelmann J, Krumbach K, Noack S, Bott M, Marienhagen J (2016) Identification of the phd gene cluster responsible for phenylpropanoid utilization in Corynebacterium glutamicum. Appl Microbiol Biotechnol 100:1871–1881. https://doi.org/10.1007/s00253-015-7165-1
Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang X-J, Zhao Y (2006b) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23:607–618. https://doi.org/10.1016/j.molcel.2006.06.026
Kim YH, Cho K, Yun S-H, Kim JY, Kwon K-H, Yoo JS, Kim SI (2006a) Analysis of aromatic catabolic pathways in Pseudomonas putida KT 2440 using a combined proteomic approach: 2-DE/MS and cleavable isotope-coded affinity tag analysis. Proteomics 6:1301–1318. https://doi.org/10.1002/pmic.200500329
Kim I-K, Yim H-S, Kim M-K, Kim D-W, Kim Y-M, Cha S-S, Kang S-O (2008) Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli. J Mol Biol 379:372–384. https://doi.org/10.1016/j.jmb.2008.04.003
Konishi Y, Shimizu M (2003) Transepithelial transport of ferulic acid by monocarboxylic acid transporter in Caco-2 cell monolayers. Biosci Biotechnol Biochem 67:856–862. https://doi.org/10.1271/bbb.67.856
Kornberg HL, Krebs HA (1957) Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988–991. https://doi.org/10.1038/179988a0
Krings U, Pilawa S, Theobald C, Berger RG (2001) Phenyl propenoic side chain degradation of ferulic acid by Pycnoporus cinnabarinus—elucidation of metabolic pathways using [5-2H]-ferulic acid. J Biotechnol 85:305–314. https://doi.org/10.1016/S0168-1656(00)00396-5
Larroy C, Fernández MR, González E, Parés X, Biosca JA (2002) Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem J 361:163–172. https://doi.org/10.1042/0264-6021:3610163
Lee E-G, Yoon S-H, Das A, Lee S-H, Li C, Kim J-Y, Choi M-S, Oh D-K, Kim S-W (2009) Directing vanillin production from ferulic acid by increased acetyl-CoA consumption in recombinant Escherichia coli. Biotechnol Bioeng 102:200–208. https://doi.org/10.1002/bit.22040
Li K, Frost JW (1998) Synthesis of vanillin from glucose. J Am Chem Soc 120:10545–10546. https://doi.org/10.1021/ja9817747
Magrane M, The UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011:bar009. https://doi.org/10.1093/database/bar009
Meinert C, Brandt U, Heine V, Beyert J, Schmidl S, Wübbeler JH, Voigt B, Riedel K, Steinbüchel A (2017) Proteomic analysis of organic sulfur compound utilisation in Advenella mimigardefordensis strain DPN7T. PLoS One 12:1–26. https://doi.org/10.1371/journal.pone.0174256
Merkens H, Beckers G, Wirtz A, Burkovski A (2005) Vanillate metabolism in Corynebacterium glutamicum. Curr Microbiol 51:59–65. https://doi.org/10.1007/s00284-005-4531-8
Meyer F, Pupkes H, Steinbüchel A (2017) Development of an improved system for the generation of knockout mutants of Amycolatopsis sp. strain ATCC 39116. Appl Environ Microbiol 83:e02660–e02616. https://doi.org/10.1128/AEM.02660-16
Mitra A, Kitamura Y, Gasson MJ, Narbad A, Parr AJ, Payne J, Rhodes MJC, Sewter C, Walton NJ (1999) 4-Hydroxycinnamoyl-CoA hydratase/lyase (HCHL)—an enzyme of phenylpropanoid chain cleavage from Pseudomonas. Arch Biochem Biophys 365:10–16. https://doi.org/10.1006/abbi.1999.1140
Monks TJ, Hanzlik RP, Cohen GM, Ross D, Graham DG (1992) Quinone chemistry and toxicity. Toxicol Appl Pharmacol 112:2–16. https://doi.org/10.1016/0041-008X(92)90273-U
Muheim A, Lerch K (1999) Towards a high-yield bioconversion of ferulic acid to vanillin. Appl Microbiol Biotechnol 51:456–461. https://doi.org/10.1007/s002530051416
Nadkarni SR, Patel MV, Chatterjee S, Vijayakumar EKS, Desikan KR, Blumbach J, Ganguli BN, Limbert M (1994) Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. Taxonomy, production, isolation and biological activity. J Antibiot (Tokyo) 47:334–341. https://doi.org/10.7164/antibiotics.47.334
Otani H, Lee Y-E, Casabon I, Eltis LD (2014) Characterization of p-hydroxycinnamate catabolism in a soil actinobacterium. J Bacteriol 196:4293–4303. https://doi.org/10.1128/JB.02247-14
Overhage J, Priefert H, Steinbüchel A (1999) Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Env Microbiol 65:4837–4847
Pfennig N (1974) Rhodopseudomonas globiformis, sp. n., a new species of the Rhodospirillaceae. Arch Microbiol 100:197–206. https://doi.org/10.1007/BF00446317
Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet 11:217–218. https://doi.org/10.1016/S0168-9525(00)89052-6
Priefert H, Rabenhorst J, Steinbüchel A (2001) Biotechnological production of vanillin. Appl Microbiol Biotechnol 56:296–314. https://doi.org/10.1007/s002530100687
Priefert H, Achterholt S, Steinbüchel A (2002) Transformation of the Pseudonocardiaceae Amycolatopsis sp. strain HR167 is highly dependent on the physiological state of the cells. Appl Microbiol Biotechnol 58:454–460. https://doi.org/10.1007/s00253-001-0920-5
Qi S-W, Chaudhry MT, Zhang Y, Meng B, Huang Y, Zhao K-X, Poetsch A, Jiang C-Y, Liu S, Liu S-J (2007) Comparative proteomes of Corynebacterium glutamicum grown on aromatic compounds revealed novel proteins involved in aromatic degradation and a clear link between aromatic catabolism and gluconeogenesis via fructose-1,6-bisphosphatase. Proteomics 7:3775–3787. https://doi.org/10.1002/pmic.200700481
Rabenhorst J, Hopp R (1997) Verfahren zur Herstellung von Vanillin und dafür geeignete Mikroorganismen. DE 195 32 317 A1. Germany
Raberg M, Reinecke F, Reichelt R, Malkus U, König S, Pötter M, Fricke WF, Pohlmann A, Voigt B, Hecker M, Friedrich B, Bowien B, Steinbüchel A (2008) Ralstonia eutropha H16 flagellation changes according to nutrient supply and state of poly(3-hydroxybutyrate) accumulation. Appl Environ Microbiol 74:4477–4490. https://doi.org/10.1128/AEM.00440-08
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. https://doi.org/10.1093/bioinformatics/16.10.944
Salmon RC, Cliff MJ, Rafferty JB, Kelly DJ (2013) The CouPSTU and TarPQM transporters in Rhodopseudomonas palustris: redundant, promiscuous uptake systems for lignin-derived aromatic substrates. PLoS One 8:1–13. https://doi.org/10.1371/journal.pone.0059844
Schlegel HG, Kaltwasser H, Gottschalk G (1961) Ein Submersverfahren zur Kultur wasserstoffoxydierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Mikrobiol 38:209–222. https://doi.org/10.1007/BF00422356
Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K (1996) Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178:7276–7284. https://doi.org/10.1128/jb.178.24.7276-7284.1996
Shimizu M, Kobayashi Y, Tanaka H, Wariishi H (2005) Transportation mechanism for vanillin uptake through fungal plasma membrane. Appl Microbiol Biotechnol 68:673–679. https://doi.org/10.1007/s00253-005-1933-2
Simon O, Klaiber I, Huber A, Pfannstiel J (2014) Comprehensive proteome analysis of the response of Pseudomonas putida KT2440 to the flavor compound vanillin. J Proteome 109:212–227. https://doi.org/10.1016/j.jprot.2014.07.006
Sutherland JB, Crawford DL, Pometto AL (1983) Metabolism of cinnamic, p-coumaric, and ferulic acids by Streptomyces setonii. Can J Microbiol 29:1253–1257. https://doi.org/10.1139/m83-195
Tyler B (1978) Regulation of the assimilation of nitrogen compounds. Annu Rev Biochem 47:1127–1162. https://doi.org/10.1146/annurev.bi.47.070178.005403
Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, Engelmann S (2008) Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8:3139–3153. https://doi.org/10.1002/pmic.200701062a
Acknowledgements
We thank Isabelle Plugge for excellent technical assistance. We also thank Robin Vivod and Kim Cohrs for helpful discussions and proofreading.
Funding
This paper was financially supported by King Abdulaziz University under Grant no. 2-10-1432/HiCi. The project was also partially funded by SYMRISE AG, Holzminden, Germany. SYMRISE AG had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 513 kb)
Rights and permissions
About this article
Cite this article
Meyer, F., Netzer, J., Meinert, C. et al. A proteomic analysis of ferulic acid metabolism in Amycolatopsis sp. ATCC 39116. Appl Microbiol Biotechnol 102, 6119–6142 (2018). https://doi.org/10.1007/s00253-018-9061-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9061-y