Abstract
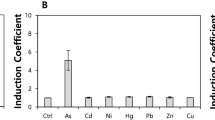
The key component in bacteria-based biosensors is a transcriptional reporter employed to monitor induction or repression of a reporter gene corresponding to environmental change. In this study, we made a series of reporters in order to achieve highly sensitive detection of arsenite. From these reporters, two biosensors were developed by transformation of Escherichia coli DH5α with pLHPars9 and pLLPars9, consisting of either a high or low copy number plasmid, along with common elements of ArsR-luciferase fusion and addition of two binding sequences, one each from E. coli and Acidithiobacillus ferrooxidans chromosome, in front of the R773 ArsR operon. Both of them were highly sensitive to arsenite, with a low detection limit of 0.04 μM arsenite (~ 5 μg/L). They showed a wide dynamic range of detection up to 50 μM using high copy number pLHPars9 and 100 μM using low copy number pLLPars9. Significantly, they differ in metal specificity, pLLPars9 more specific to arsenite, while pLHPars9 to both arsenite and antimonite. The only difference between pLHPars9 and pLLPars9 is their copy numbers of plasmid and corresponding ratios of ArsR to its binding promoter/operator sequence. Therefore, we propose a working model in which DNA bound-ArsR is different from its free form in metal specificity.






Similar content being viewed by others
References
Ambriz-Aviña V, Contreras-Garduño JA, Pedraza-Reyes M (2014) Applications of flow cytometry to characterize bacterial physiological responses. Biomed Res Int 2014:1–14
Busenlehner LS, Pennella MA, Giedroc DP (2003) The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev 27:131–143
Butcher BG, Rawlings DE (2002) The divergent chromosomal ars operon of Acidithiobacillus ferrooxidans is regulated by an atypical ArsR protein. Microbiology 148:3983–3992
Chen J, Rosen BP (2014) Biosensors for inorganic and organic arsenicals. Biosensors 4:494–512
Chen J, Zhu YG, Rosen BP (2012) A novel biosensor selective for organoarsenicals. Appl Environ Microbiol 78:7145–7147
Chu J (2007) A safe and simple arsenic detector. MIT Technololy Review. https://www.technologyreview.com/s/407222/a-safe-and-simple-arsenic-detector/
Close DM, Ripp S, Sayler GS (2009) Reporter proteins in whole-cell optical bioreporter detection systems, biosensor integrations, and biosensing applications. Sensors 9:9147–9174
Fernández M, Morel B, Ramos JL, Krell T (2016) Paralogous regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a basis for arsenic biosensor development. Appl Environ Microbiol 82:4133–4144
Gilman J, Love J (2016) Synthetic promoter design for new microbial chassis. Biochem Soc Trans 44:731–737
Hödar C, Moreno P, Genova A, Latorre M, Reyes-Jara A, Maass A, González M, Cambiazo V (2012) Genome wide identification of Acidithiobacillus ferrooxidans (ATCC 23270) transcription factors and comparative analysis of ArsR and MerR metal regulators. Biometals 25:75–93
Hughes MF, Beck BD, Chen Y, Lewis AS, Thomas DJ (2011) Arsenic exposure and toxicology: a historical perspective. Toxicol Sci 123:305–332
Jomova K, Jenisova Z, Feszterova M, Baros S, Liska J, Hudecova D, Rhodes CJ, Valko M (2011) Arsenic: toxicity, oxidative stress and human disease. J Appl Toxicol 31:95–107
Juers DH, Matthews BW, Huber RE (2012) LacZ, β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21:1792–1807
Kaur H, Kumar R, Babu JN, Mittal S (2015) Advances in arsenic biosensor development—a comprehensive review. Biosens Bioelectron 63:533–545
Lee PE, Demple B, Barton JK (2009) DNA-mediated redox signaling for transcriptional activation of SoxR. Proc Natl Acad Sci U S A 106:13164–13168
Li L, Liang J, Hong W, Zhao Y, Sun S, Yang X, Xu A, Hang H, Wu L, Chen S (2015) Evolved bacterial biosensor for arsenite detection in environmental water. Environ Sci Technol 49:6149–6155
McElroy WD, Seliger HH (1963) The chemistry of light emission. Adv Enzymol Relat Areas Mol Biol John Wiley & Sons, Inc. (25):119–166
Meighen EA (1988) Enzymes and genes from the lux operon of bioluminescent bacteria. Annu Rev Microbiol 42:151–176
Merulla D, van der Meer JR (2016) Regulatable and modulable background expression control in prokaryotic synthetic circuits by auxiliary repressor binding sites. ACS Synth Biol 5:36−45
O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG (1989) The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell 56:119–129
Outten CE, Outten FW, O’Halloran TV (1999) DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem 274:37517–37524
Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP (2007) Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem 282:34346–34355
San Francisco MJ, Hope CL, Owolabi JB, Tisa LS, Rosen BP (1990) Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res 18:619–624
Shi W, Wu J, Rosen BP (1994) Identification of a putative metal binding site in a new family of metalloregulatory proteins. J Biol Chem 269:19826–19829
Soangra R, Majumder CB, Roy P (2015) Whole cell arsenic biosensor—a cheap technology for bioavailable arsenic (As) determination. Eur J Adv Eng Technol 2:52–61
Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, Daunert S, Malik KA, van der Meer JR (2003) The development of a set of bacterial biosensors based on a nonpathogenic laboratory strain of Escherichia coli. Environ Sci Technol 37:4743–4750
Tauriainen S, Karp M, Chang W, Virta M (1997) Recombinant luminescent bacteria for measuring bioavailable arsenite and antimonite. Appl Environ Microbiol 63:4456–4461
Trang PT, Berg M, Viet PH, Mui NV, van Der Meer JR (2005) Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ Sci Technol 39:7625–7630
Wu J, Rosen BP (1993) Metalloregulated expression of the ars operon. J Biol Chem 268:52–58
Xu C, Shi W, Rosen BP (1996) The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem 271:2427–2432
Funding
This work was supported by the High-level Leading Talent Introduction Program of GDAS (2016GDASRC-0208) and the Science and Technology Planning Project of Guangzhou City (201707020021) to XL, the National Natural Science Foundation of China (31600077) and the China Postdoctoral Science Foundation Grant (2017M612622) to YF, and the Science and Technology Project of Guangdong Province (2016B070701017) to MX.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Rights and permissions
About this article
Cite this article
Fang, Y., Zhu, C., Chen, X. et al. Copy number of ArsR reporter plasmid determines its arsenite response and metal specificity. Appl Microbiol Biotechnol 102, 5753–5761 (2018). https://doi.org/10.1007/s00253-018-9042-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9042-1