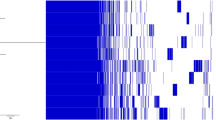
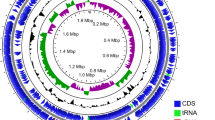
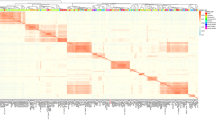
Abstract
Enterococcus spp. are present in the native microbiota of many traditional fermented foods. Their ability to produce antibacterial compounds, mainly against Listeria monocytogenes, has raised interest recently. However, there is scarce information about their proteolytic and lipolytic potential, and their biotechnological application is currently limited because enterococcal strains have been related to nosocomial infections. In this work, next-generation sequencing and optimised bioinformatic pipelines were used to annotate the genomes of two Enterococcus strains—one E. faecium and one E. faecalis—isolated from the Mexican artisanal ripened Cotija cheese. A battery of genes involved in their proteolytic system was annotated. Genes coding for lipases, esterases and other enzymes whose final products contribute to cheese aroma and flavour were identified as well. As for the production of antibacterial compounds, several peptidoglycan hydrolase- and bacteriocin-coding genes were identified in both genomes experimentally and by bioinformatic analyses. E. faecalis showed resistance to aminoglycosides and E. faecium to aminoglycosides and macrolides, as predicted by the genome functional annotation. No pathogenicity islands were found in any of the strains, although traits such as the ability of biofilm formation and cell aggregation were observed. Finally, a comparative genomic analysis was able to discriminate between the food strains isolated and nosocomial strains. In summary, pathogenic strains are resistant to a wide range of antibiotics and contain virulence factors that cause host damage; in contrast, food strains display less antibiotic resistance, include genes that encode class II bacteriocins and express virulence factors associated with host colonisation rather than invasion.






Similar content being viewed by others
References
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12(1):402. https://doi.org/10.1186/1471-2164-12-402
Alvarez-Cisneros YM, Fernández FJ, Wacher-Rodarte C, Aguilar MB, Sáinz Espuñes T del R, Ponce-Alquicira E (2010) Biochemical characterization of a bacteriocin-like inhibitory substance produced by Enterococcus faecium MXVK29, isolated from Mexican traditional sausage. J Sci Food Agric 90:2475–2481. doi: https://doi.org/10.1002/jsfa.4109, 14
Andrews S (2016) Babraham Bieoinformatics e FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 9 Sep 2016
Aziz RK, Bartels D, Best AA, De Jongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9(1):75. https://doi.org/10.1186/1471-2164-9-75
Bender JK, Fiedler S, Klare I, Werner G (2015) Complete genome sequence of the gut commensal and laboratory strain Enterococcus faecium 64/3. Genome Announc 3(6):e01275–e01215. https://doi.org/10.1128/genomeA.01275-15
Besemer J, Lomsadze A, Borodovsky M (2001) Prokaryotic GeneMark.Hmm version 2 GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618. https://doi.org/10.1093/nar/29.12.2607
Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. https://doi.org/10.1093/bioinformatics/btq683
Bourgogne A, Garsin D (2008) Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9(7):R110. https://doi.org/10.1186/gb-2008-9-7-r110
Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH (2012) A reference-free algorithm for computational normalization of shotgun sequencing data. http://arxiv.org/abs/1203.4802. Accessed 13 April 2016; arXiv:1203.4802
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinf 10(1):421. https://doi.org/10.1186/1471-2105-10-421
Carattoli A, Zankari E, Garcia-Fernandez A, Volby Larsen M, Lund O, Villa L, Aarestrup FM, Hasman H (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58(7):3895–3903. https://doi.org/10.1128/AAC.02412-14
Cebrián R, Baños A, Valdivia E, Pérez-Pulido R, Martínez-Bueno M, Maqueda M (2012) Characterization of functional, safety, and probiotic properties of Enterococcus faecalis UGRA10, a new AS-48-producer strain. Food Microbiol 30(1):59–67. https://doi.org/10.1016/j.fm.2011.12.002
Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium, Detter JC (2009) Genomics. Genome project standards in a new era of sequencing. Science 326(5950):236–237. https://doi.org/10.1126/science.1180614
Clinical and Laboratory Standards Institute (2012) Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne
Contreras-Moreira B, Vinuesa P (2013) GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79(24):7696–7701. https://doi.org/10.1128/AEM.02411-13
Dag Anders B, Lars G (2011) Complete genome sequence of the commensal Enterococcus faecalis 62, isolated from a healthy Norwegian infant. J Bacteriol 193(9):2377–2378. https://doi.org/10.1128/JB.00183-11
Darling AE, Mau B, Perna NT (2010) Progressive mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. https://doi.org/10.1371/journal.pone.0011147
Domann E, Hain T, Ghai R, Billion A, Kuenne C, Zimmermann K, Chakraborty T (2007) Comparative genomic analysis for the presence of potential enterococcal virulence factors in the probiotic Enterococcus faecalis strain Symbioflor 1. Int J Med Microbiol 297(7-8):533–539. https://doi.org/10.1016/j.ijmm.2007.02.008
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7(10):e1002195. https://doi.org/10.1371/journal.pcbi.1002195
Engels WJM, Dekker R, de Jong C, Neeter R, Visser S (1997) A comparative study of volatile compounds in the water-soluble fraction of various types of ripened cheese. Int Dairy J 7(4):255–263. https://doi.org/10.1016/S0958-6946(97)00003-4
Escobar-Zepeda A, Sanchez-Flores A, Quirasco Baruch M (2016) Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol 57:116–127. https://doi.org/10.1016/j.fm.2016.02.004
Foulquié-Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106(1):1–24. https://doi.org/10.1016/j.ijfoodmicro.2005.06.026
Franz CM, Muscholl-Silberhorn AB, Yousif NMK, Vancanneyt M, Swings J, Holzapfel WH (2001) Incidence of virulent factors and antibiotic resistance among enterococci isolated from food. Appl Environ Microbiol 67(9):4385–4389. https://doi.org/10.1128/AEM.67.9.4385-4389.2001
Franz CM, van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A (2007) Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev 31(3):293–310. https://doi.org/10.1111/j.1574-6976.2007.00064.x
Franz CM, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011) Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151(2):125–140. https://doi.org/10.1016/j.ijfoodmicro.2011.08.014
Frees D, Savijoki K, Varmanen P, Ingmer H (2007) Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, gram-positive bacteria. Mol Microbiol 63(5):1285–1295. https://doi.org/10.1111/j.1365-2958.2007.05598.x
Fritzenwanker M, Kuenne C (2013) Complete genome sequence of the probiotic Enterococcus faecalis Symbioflor 1 clone DSM 16431. Genome Announc 1(1):e00165–e00112. https://doi.org/10.1128/genomeA.00165-12
García-Cano I, Serrano-Maldonado CE, Olvera-García M, Delgado Arciniega E, Peña-Montes C, Quirasco M (2014) Antibacterial activity produced by Enterococcus spp. isolated from artisanal Mexican dairy product, Cotija cheese. LWT Food Sci Technol 59(1):26–34. https://doi.org/10.1016/j.lwt.2014.04.059
Garsin DA, Frank KL, Silanpää J, Ausubel FM, Hartke A, Shankar N, Murray BE (2014) Pathogenesis and models of enterococcal infection. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (eds) Enterococci: from commensals to leading causes of drug resistant infection [internet]. Eye and Ear Infirmary, Boston, pp 135–169
Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35(Web Server):W52–W57. https://doi.org/10.1093/nar/gkm360
Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD (2013) REAPR: a universal tool for genome assembly evaluation. Genome Biol 14(5):R47. https://doi.org/10.1186/gb-2013-14-5-r47
Jerse AE, Yu J, Tall BD, Kaper JB (1990) A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87(20):7839–7843. https://doi.org/10.1073/pnas.87.20.7839
Kim EB, Marco ML (2014) Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl Environ Microbiol 80(1):154–165. https://doi.org/10.1128/AEM.03108-13
Kopit LM, Kim EB, Siezen RJ, Harris LJ, Marco ML (2014) Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Appl Environ Microbiol 80(6):1899–1909. https://doi.org/10.1128/AEM.03859-13
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. https://doi.org/10.1006/jmbi.2000.4315
Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP (2012) Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 194(9):2334–2341. https://doi.org/10.1128/JB.00259-12
Lam MM, Seeman T, Tobias NJ, Chen H, Haring V, Moore RJ, Ballard S, Grayson LM, Johnson PD, Howden BP, Stinear TP (2013) Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 14(1):595. https://doi.org/10.1186/1471-2164-14-595
Lamarque M, Charbonnel P, Aubel D, Piard JC, Atlan D, Juillard V (2004) A multifunction ABC transporter (opt) contributes to diversity of peptide uptake specificity within the genus Lactococcus. J Bacteriol 186(19):6492–6500. https://doi.org/10.1128/JB.186.19.6492-6500.2004
Lepeuple AS, van Gemert E, Chapot-Chartier MP (1998) Analysis of the bacteriolytic enzymes of the autolytic Lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl Environ Microbiol 64:4142–4148
Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ (2010) The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genomics 11(1):36. https://doi.org/10.1186/1471-2164-11-36
Mesnage S, Chau F, Dubost L, Arthur M (2008) Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J Biol Chem 283(28):19845–19853. https://doi.org/10.1074/jbc.M802323200
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35(Web Server):W182–W185. https://doi.org/10.1093/nar/gkm321
Nadalin F, Vezzi F, Policriti A (2012) GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinf 14(Suppl 14):S8. https://doi.org/10.1186/1471-2105-13-S14-S8
Natarajan P, Parani M (2015) First complete genome sequence of a probiotic Enterococcus faecium strain T-110 and its comparative genome analysis with pathogenic and non-pathogenic Enterococcus faecium genomes. J Genet Genomics 42(1):43–46. https://doi.org/10.1016/j.jgg.2014.07.002
Navarro-García F, Eslava C, Villaseca JM, López-Revilla R, Czeczulin JR, Srinivas S, Nataro JP, Cravioto A (1998) In vitro effects of a high-molecular-weight heat-labile enterotoxin from enteroaggregative Escherichia coli. Infect Immun 66(7):3149–3154
Nes IF, Diep DB, Ike Y (2014) Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (eds) Enterococci: from commensals to leading causes of drug resistant infection [internet]. Eye and Ear Infirmary, Boston, pp 446–482
NMX-F-735-COFOCALEC (2011) Mexican Official Standard. Sistema producto leche-alimentos-lácteos-alimento lácteo regional-queso Cotija artesanal madurado-denominación, especificaciones y métodos de prueba. DOF: 24/06/2011, Diario Oficial de la Federación, Mexico City
Nueno-Palop C, Narbad A (2011) Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int J Food Microbiol 145(2-3):390–394. https://doi.org/10.1016/j.ijfoodmicro.2010.12.029
Orgier JC, Serror P (2008) Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126(3):291–301. https://doi.org/10.1016/j.ijfoodmicro.2007.08.017
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42(D1):D206–D214. https://doi.org/10.1093/nar/gkt1226
Palmer KL, Gilmore MS (2010) Multidrug-resistant entorococci lack CRISPR-cas. mBio 1(4):e00227-10. https://doi.org/10.1128/mBio.00227-10
Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS (2012) Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. https://doi.org/10.1128/mBio.00318-11
Palmer KL, van Schaik W, Willems RJL, Gilmore MS (2014) Enterococcal genomics. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (eds) Enterococci: from commensals to leading causes of drug resistant infection [internet]. Eye and Ear Infirmary, Boston, pp 189–216
Park SS, Kwon HY, Tran TD, Choi MH, Jung SH, Lee S, Briles DE, Rhee DK (2015) ClpL is a chaperone without auxiliary factors. FEBS J 282(8):1352–1367. https://doi.org/10.1111/febs.13228
Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299(5615):2071–2074. https://doi.org/10.1126/science.1080613
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786. https://doi.org/10.1038/nmeth.1701
Ponstingl H, Ning Z (2010) SMALT. http://www.sanger.ac.uk/science/tools/smalt-0. Accessed 13 April 2016
Qin X, Galloway-Peña JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny DM, Buhay CJ, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino JF, Worley KC, Gibbs RA, Weinstock GM, Murray BE (2012) Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12(1):135. https://doi.org/10.1186/1471-2180-12-135
Sarantinopoulos P, Andrighetto C, Georgalakia MD, Rea MC, Lombardi A, Coganc TM, Kalantzopoulosa G, Tsakalidou E (2001) Biochemical properties of enterococci relevant to their technological performance. Int Dairy J 11(8):621–647. https://doi.org/10.1016/S0958-6946(01)00087-5
Sarantinopoulos P, Kalantzopoulos G, Tsakalidou E (2002) Effect of Enterococcus faecium on microbiological, physicochemical and sensory characteristics of Greek feta cheese. Int J Food Microbiol 76(1-2):93–105. https://doi.org/10.1016/S0168-1605(02)00021-1
Savijoki K, Ingmer H, Varmanen P (2006) Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol 71(4):394–406. https://doi.org/10.1007/s00253-006-0427-1
Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in range from 1 to 100 kDa. Anal Biochem 166(2):368–379. https://doi.org/10.1016/0003-2697(87)90587-2
Serio A, Chaves-López C, Paparella A, Suzzi G (2010) Evaluation of metabolic activities of enterococci isolated from pecorino Abruzzese cheese. Int Dairy J 20(7):459–464. https://doi.org/10.1016/j.idairyj.2010.02.005
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19(6):1117–1123. https://doi.org/10.1101/gr.089532.108
Smit G, Smit BA, Engels WJM (2005) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 29(3):591–610. https://doi.org/10.1016/j.femsre.2005.04.002
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 12(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Uchimura Y, Wyss M, Brugiroux S, Limenitakis JP, Stecher B, McCoy KD, Macpherson AJ (2016) Complete genome sequences of 12 species of stable defined moderately diverse mouse microbiota 2. Genome Announc 4(5):e00951–e00916. https://doi.org/10.1128/genomeA.00951-16
van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP (2013) BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal post translationally modified peptides. Nucleic Acids Res 41(W1):W448–W453. https://doi.org/10.1093/nar/gkt391
van Kranenburg R, Kleerebezem M, van HylckamaVlieg J, Ursing BM, Boekhorst J, Smit BA, Ayad EHE, Smit G, Siezen RJ (2002) Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int Dairy J 12(2-3):111–121. https://doi.org/10.1016/S0958-6946(01)00132-7
Weigand MR, Ashbolt NJ, Konstantinidis KT, Santo Domingo JW (2014) Genome sequencing reveals the environmental origin of enterococci and potential biomarkers for water quality monitoring. Environ Sci Technol 48(7):3707–3714. https://doi.org/10.1021/es4054835
Wunderlichová L, Bunková L, Koutný M, Jancová P, Bunka F (2014) Formation, degradation, and detoxification of putrescine by foodborne bacteria: a review. Compr Rev Food Sci Food Saf 13(5):1012–1030. https://doi.org/10.1111/1541-4337.12099
Yoon SH, Park YK, Kim JF (2014) PAIDB v2.0: exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res 43(D1):D624–D630. https://doi.org/10.1093/nar/gku985
Yuehua J, Lanwei Z, Fei L, Huaxi Y, Xue H (2016) Complete genome sequence of Enterococcus faecalis LD33, a bacteriocin-producing strain. J Biotechnol 227:79–80. https://doi.org/10.1016/j.jbiotec.2016.04.030
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Zischka M, Kuenne C (2012) Complete genome sequence of the porcine isolate Enterococcus faecalis D32. J Bacteriol 194(19):5490–5491. https://doi.org/10.1128/JB.01298-12
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acid Res 39(suppl):W347–W352. https://doi.org/10.1093/nar/gkr485
Acknowledgments
This work was supported by UNAM-PAPIIT IN222115 and IN222717 grants. We thank Alejandra Escobar-Zepeda, Jérôme Verleyen and Veronica Jiménez-Jacinto for their support in bioinformatic analyses, which were performed in the ‘Unidad de Secuenciación Masiva y Bioinformática’ at the ‘Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas’, CONACyT #260481, Instituto de Biotecnología/UNAM; to Dr. Edith Ponce Alquicira, from Universidad Autónoma Metropolitana-Iztapalapa (UAM-I) for providing the Enterococcus faecium strain MXVK29 and Enterococcus faecalis NCIMB-700585; and to Dr. Carlos Eslava-Campos, for providing mammal cell lines and for his assistance in performing the adherence experiments. M.O.-G. was granted a doctoral fellowship from Consejo Nacional de Ciencia y Tecnología (CONACyT). We also thank Dr. Romina Rodríguez Sanoja and Dr. Rosa María Gutierrez for their valuable comments and support during the development of this project. María Elena Sánchez-Salazar edited the English manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statement
This manuscript is in compliance with ethical standards. This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 54 kb)
Rights and permissions
About this article
Cite this article
Olvera-García, M., Sanchez-Flores, A. & Quirasco Baruch, M. Genomic and functional characterisation of two Enterococcus strains isolated from Cotija cheese and their potential role in ripening. Appl Microbiol Biotechnol 102, 2251–2267 (2018). https://doi.org/10.1007/s00253-018-8765-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-8765-3