Abstract
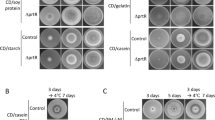
Three extracellular dipeptidyl peptidase genes, dppB, dppE, and dppF, were unveiled by sequence analysis of the Aspergillus oryzae genome. We investigated their differential enzymatic profiles, in order to gain an understanding of the diversity of these genes. The three dipeptidyl peptidases were expressed using Aspergillus nidulans as the host. Each recombinant enzyme was purified and subsequently characterized. The enzymes displayed similar optimum pH values, but optimum temperatures, pH stabilities, and substrate specificities varied. DppB was identified as a Xaa-Prolyl dipeptidyl peptidase, while DppE scissile substrates were similar to the substrates for Aspergillus fumigatus DPPV (AfDPPV). DppF was found to be a novel enzyme that could digest both substrates for A. fumigatus DPPIV and AfDPPV. Semi-quantitative PCR revealed that the transcription of dppB in A. oryzae was induced by protein substrates and repressed by the addition of an inorganic nitrogen source, despite the presence of protein substrates. The transcription of dppE depended on its growth time, while the transcription of dppF was not affected by the type of the nitrogen source in the medium, and it started during the early stage of the fungal growth. Based on these results, we conclude that these enzymes may represent the nutrition acquisition enzymes. Additionally, DppF may be one of the sensor peptidases responsible for the detection of the protein substrates in A. oryzae environment. DppB may be involved in nitrogen assimilation control, since the transcription of dppB was repressed by NaNO3, despite the presence of protein substrates.




Similar content being viewed by others
References
Beauvais A, Monod M, Debeaupuis JP, Diaquin M, Kobayashi H, Latgé JP (1997a) Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J Biol Chem 272:6238–6244. doi:10.1074/jbc.272.10.6238
Beauvais A, Monod M, Wyniger J, Debeaupuis JP, Grouzmann E, Brakch N, Svab J, Hovanessian AG, Latgé JP (1997b) Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect Immun 65:3042–3047
Bergmann A, Hartmann T, Cairns T, Bignell EM, Krappmann S (2009) A regulator of Aspergillus fumigatus extracellular proteolytic activity is dispensable for virulence. Infect Immun 77:4041–4050. doi:10.1128/IAI.00425-09
Doumas A, van den Broek P, Affolter M, Monod M (1998) Characterization of the prolyl dipeptidyl peptidase gene (dppIV) from the koji mold Aspergillus oryzae. Appl Environ Microbiol 64:4809–4815
Fujioka T, Mizutani O, Furukawa K, Sato N, Yoshimi A, Yamagata Y, Nakajima T, Abe K (2007) MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot Cell 6:1497–1510. doi:10.1128/EC.00281-06
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. doi:10.1038/nature04341
Gomi K, Iimura Y, Hara S (1987) Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric Biol Chem 51:2549–2555. doi:10.1271/bbb1961.51.2549
Haltmann T, Cairns TC, Olbermann P, Morschhäuser J, Bignell EM, Krappmann S (2011) Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol Microbiol 82:917–935. doi:10.1111/j.1365-2958.2011.07868.x
Ichishima E (2000) Unique catalytic and molecular properties of hydrolases from Aspergillus used in Japanese bioindustries. Biosci Biotechnol Biochem 64:675–688. doi:10.1271/bbb.64.675
Ishida K, Kuboshima M, Morita H, Maeda H, Okamoto A, Takeuchi M, Yamagata Y (2014) Diversity in mRNA expression of the serine-type carboxypeptidase ocpG in Aspergillus oryzae through intron retention. Biosci Biotechnol Biochem 78:1328–1336. doi:10.1080/09168451.2014.923291
Kitamoto K (2002) Molecular biology of the Koji molds. Adv Appl Microbiol 51:129–153. doi:10.1016/S0065-2164(02)51004-2
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi:10.1038/227680a0
Machida M (2002) Progress of Aspergillus oryzae genomics. Adv Appl Microbiol 51:81–106. doi:10.1016/S0065-2164(02)51002-9
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, DW Denning, JE Galagan, WC Nierman, Yu J, DB Archer, JW Bennett, D Bhatnagar, TE Cleveland, ND Fedorova, O Gotoh, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, PR Juvvadi, M Kato, Y Kato, T Kin, A Kokubun, H Maeda, N Maeyama, J Maruyama, H Nagasaki, T Nakajima, K Oda, K Okada, I Paulsen, K Sakamoto, T Sawano, M Takahashi, K Takase, Y Terabayashi, JR Wortman, O Yamada, Y Yamagata, H Anazawa, Y Hata, Y Koide, T Komori, Y Koyama, T Minetoki, S Suharnan, A Tanaka, K Isono, S Kuhara, N Ogasawara, H Kikuchi (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. doi:10.1038/nature04300
Maley F, Trimble RB, Tarentino AL, Plummer TH Jr (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem 180:195–204. doi:10.1016/0003-2697(89)90115-2
Matsudaira P (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 262:10035–10038
Minetoki T, Tsuboi H, Koda A, Ozeki K (2003) Development of high expression system with the improved promoter using the cis-acting elements in Aspergillus species. J Biol Macromol 3:89–96
Monod M, Beauvais A (2004) Dipeptidyl-peptidases IV and V of Aspergillus. In: Barrett AJ, Rawlings ND, Woessner JF (eds) Handbook of proteolytic enzymes, vol 2, 2nd edn. Elsevier, London, pp 1911–1913
Morita H, Kuriyama K, Akiyama N, Okamoto A, Yamagata Y, Kusumoto K-I, Koide Y, Ishida H, Takeuchi M (2010) Molecular cloning of ocpO encoding carboxypeptidase O of Aspergillus oryzae IAM2640. Biosci Biotechnol Biochem 74:1000–1006
Motoyama T, Kojima N, Horiuchi H, Ohta A, Takagi M (1994) Isolation of a chitin synthase gene (chsC) of Aspergillus nidulans. Biosci Biotechnol Biochem 58:2254–2257. doi:10.1271/bbb.58.2254
Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O’Neil S, Paulsen I, Peñalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Córdoba S, Rodríguez-Peña JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sánchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi:10.1038/nature04332
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wösten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. doi:10.1038/nbt1282
Sambrook J, Fritsh EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY
Schechter I, Berger A (1967) On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27:157–162
Tachi H, Ito H, Ichishhima E (1992) An X-proly dipeptidyl peptidase from Aspergillus oryzae. Phytochemistry 31:3707–3709. doi:10.1016/S0031-9422(00)97513-7
Taylor MJ, Richardson T (1979) Applications of microbial enzymes in food systems and in biotechnology. Adv Appl Microbiol 25:7–35. doi:10.1016/S0065-2164(08)70144-8
Tudzynski B (2014) Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol 28:656. doi:10.3389/fmicb.2014.00656
Wiechelman KJ, Braun RD, Fitzpatrick JD (1988) Investigation of the bicinchoninic acid protein assay: identification of the groups responsible for color formation. Anal Biochem 175:231–237. doi:10.1016/0003-2697(88)90383-1
Wolfinbarger L Jr, Marzluf GA (1974) Peptide utilization by amino acid auxotrophs of Neurospora crassa. J Bacteriol 119:371–378
Yamamoto S, Shiga K, Kodama Y, Imamura M, Uchida R, Obata A, Bamba T, Fukusaki E (2014) Analysis of the correlation between dipeptides and taste differences among soy sauces by using metabolomics-based component profiling. J Biosci Bioeng 118:56–63. doi:10.1016/j.jbiosc.2013.12.019
Yike I (2011) Fungal proteases and their pathophysiological effects. Mycopathologia 171:299–323. doi:10.1007/s11046-010-9386-2
Acknowledgments
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any study with human participants or animals performed by any of the authors.
Electronic Supplementary Material
ESM 1
(PDF 686 kb)
Rights and permissions
About this article
Cite this article
Maeda, H., Sakai, D., Kobayashi, T. et al. Three extracellular dipeptidyl peptidases found in Aspergillus oryzae show varying substrate specificities. Appl Microbiol Biotechnol 100, 4947–4958 (2016). https://doi.org/10.1007/s00253-016-7339-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7339-5