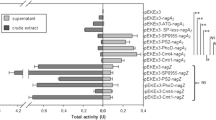
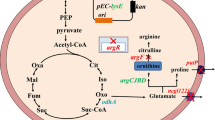
Abstract
Sustainable supply of feedstock has become a key issue in process development in microbial biotechnology. The workhorse of industrial amino acid production Corynebacterium glutamicum has been engineered towards utilization of alternative carbon sources. Utilization of the chitin-derived aminosugar N-acetyl-glucosamine (GlcNAc) for both cultivation and production with C. glutamicum has hitherto not been investigated. Albeit this organism harbors the enzymes N-acetylglucosamine-6-phosphatedeacetylase and glucosamine-6P deaminase of GlcNAc metabolism (encoded by nagA and nagB, respectively) growth of C. glutamicum with GlcNAc as substrate was not observed. This was attributed to the lack of a functional system for GlcNAc uptake. Of the 17 type strains of the genus Corynebacterium tested here for their ability to grow with GlcNAc, only Corynebacterium glycinophilum DSM45794 was able to utilize this substrate. Complementation studies with a GlcNAc-uptake deficient Escherichia coli strain revealed that C. glycinophilum possesses a nagE-encoded EII permease for GlcNAc uptake. Heterologous expression of the C. glycinophilum nagE in C. glutamicum indeed enabled uptake of GlcNAc. For efficient GlcNac utilization in C. glutamicum, improved expression of nagE with concurrent overexpression of the endogenous nagA and nagB genes was found to be necessary. Based on this strategy, C. glutamicum strains for the efficient production of the amino acid l-lysine as well as the carotenoid lycopene from GlcNAc as sole substrate were constructed.



Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Arndt A, Eikmanns BJ (2008) Regulation of carbon metabolism in Corynebacterium glutamicum. In: Burkovski A (ed) Corynebacteria: genomics and molecuar biology. Caister Acadeic Press, Norfolk, UK, pp 155–182
Becker J, Wittmann C (2012) Bio-based production of chemicals, materials and fuels —Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640
Becker J, Schafer R, Kohlstedt M, Harder BJ, Borchert NS, Stoveken N, Bremer E, Wittmann C (2013) Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb Cell Factories 12:110
Beier S, Bertilsson S (2013) Bacterial chitin degradation—mechanisms and ecophysiological strategies. Front Microbiol 4:149
Bertram R, Rigali S, Wood N, Lulko AT, Kuipers OP, Titgemeyer F (2011) Regulon of the N-acetylglucosamine utilization regulator NagR in Bacillus subtilis. J Bacteriol 193:3525–3536
Bhattacharya D, Nagpure A, Gupta RK (2007) Bacterial chitinases: properties and potential. Crit Rev Biotechnol 27:21–28
Bhattachrya D, Nagpure A, Gupta RK (2007) Bacterial chitinase: properties and potential. Crit Rev Biotechnol 27:21–28
Blombach B, Seibold GM (2010) Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol 86:1313–1322
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Bückle-Vallant V, Krause FS, Messerschmidt S, Eikmanns BJ (2014) Metabolic engineering of Corynebacterium glutamicum for 2-ketoisocaproate production. Appl Microbiol Biotechnol 98:297–311
Buschke N, Schafer R, Becker J, Wittmann C (2013) Metabolic engineering of industrial platform microorganisms for biorefinery applications—optimization of substrate spectrum and process robustness by rational and evolutive strategies. Bioresour Technol 135:544–554
Chen JK, Shen CR, Liu CL (2010) N-Acetylglucosamine: production and applications. Mar Drugs 8:2493–2516
Claes WA, Puhler A, Kalinowski J (2002) Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J Bacteriol 184:2728–2739
Comb DG, Roseman S (1956) Glucosamine-6-phosphate deaminase. Biochim Biophys Acta 21:193–194
Eggeling L, Reyes O (2005) Experiments. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC, Boca Raton, FL, pp 3535–3566
Eijsink VG, Vaaje-Kolstad G, Varum KM, Horn SJ (2008) Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol 26:228–235
Eijsink V, Hoell I, Vaaje-Kolstada G (2010) Structure and function of enzymes acting on chitin and chitosan. Biotechnol Genet Eng Rev 27:331–366
Eikmanns BJ, Thum-Schmitz N, Eggeling L, Lüdtke KU, Sahm H (1994) Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817–1828
Engels V, Wendisch VF (2007) The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J Bacteriol 189:2955–2966
Engels V, Georgi T, Wendisch VF (2008a) ScrB (Cg2927) is a sucrose-6-phosphate hydrolase essential for sucrose utilization by Corynebacterium glutamicum. FEMS Microbiol Lett 289:80–89
Engels V, Lindner SN, Wendisch VF (2008b) The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J Bacteriol 190:8033–8044
Gaigalat L, Schluter JP, Hartmann M, Mormann S, Tauch A, Puhler A, Kalinowski J (2007) The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol Biol 8:104
Georgi T, Rittmann D, Wendisch VF (2005) Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab Eng 7:291–301
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Gillett R (2008) Global study of shrimp fisheries. Food and Agriculture Organization of the United Nations, Rome
Gopinath V, Meiswinkel TM, Wendisch VF, Nampoothiri KM (2011) Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl Microbiol Biotechnol 92(5):985–996
Gruteser N, Marin K, Kramer R, Thomas GH (2012) Sialic acid utilization by the soil bacterium Corynebacterium glutamicum. FEMS Microbiol Lett 336:131–138
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580
Hayes M, Carney B, Slater J, Bruck W (2008) Mining marine shellfish wastes for bioactive molecules: chitin and chitosan—Part A. Extraction methods. Biotechnol J 3(7):871–877
Heider SA, Peters-Wendisch P, Wendisch VF (2012) Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol 12:198
Heider SA, Peters-Wendisch P, Netzer R, Stafnes M, Brautaset T, Wendisch VF (2013) Production and glucosylation of C and C carotenoids by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol (Epub ahead of print)
Inokuma K, Takano M, Hoshino K (2013) Direct ethanol production from N-acetylglucosamine and chitin substrates by Mucor species. Biochem Eng J 72:24–32
Inui M, Kawaguchi H, Murakami S, Vertes AA, Yukawa H (2004) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254
Johnson JW, Fisher JF, Mobashery S (2013) Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Kandra P, Challa MM, Jyothi HKP (2012) Efficient use of shrimp waste: present and future trends. Appl Microbiol Biotechnol 93:17–29
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, Wendisch VF (2010) Quinone-dependent d-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on d-lactate. BMC Microbiol 10:321
Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72:3418–3428
Kawaguchi H, Sasaki M, Vertes AA, Inui M, Yukawa H (2008) Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol 77:1053–1062
Krause FS, Blombach B, Eikmanns BJ (2010) Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol 76:8053–6061
Krings E, Krumbach K, Bathe B, Kelle R, Wendisch VF, Sahm H, Eggeling L (2006) Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on l-lysine formation. J Bacteriol 188:8054–8061
Kurita K (2006) Chitin and chitosan: functional biopolymers from marine crustaceans. Mar Biotechnol 8:203–826
Lengeler J (1975) Mutations affecting transport of the hexitols d-mannitol, d-glucitol, and galactitol in Escherichia coli K-12: isolation and mapping. J Bacteriol 124:26–38
Lengeler J (1980) Characterisation of mutants of Escherichia coli K12, selected by resistance to streptozotocin. Mol Gen Genet 179:49–54
Lengeler J, Auburger AM, Mayer R, Pecher A (1981) The phosphoenolpyruvate-dependent carbohydrate: phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K 12. Mol Gen Genet 183:163–170
Lindner SN, Seibold GM, Henrich A, Krämer R, Wendisch VF (2011) Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microbiol 77:3571–3581
Litsanov B, Kabus A, Brocker M, Bott M (2012) Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol 5:116–128
Martin JF, Barreiro C, Gonzalez-Lavado E, Barriuso M (2003) Ribosomal RNA and ribosomal proteins in corynebacteria. J Biotechnol 104:41–53
Meiswinkel TM, Gopinath V, Lindner SN, Nampoothiri KM, Wendisch VF (2013a) Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol 6:131–140
Meiswinkel TM, Rittmann D, Lindner SN, Wendisch VF (2013b) Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour Technol 145:254–258
Mimitsuka T, Sawai H, Hatsu M, Yamada K (2007) Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem 71:2130–2135
Nakada HI, Wolfe JB (1956) Glucosamine degradation by Escherichia coli: II. The isomeric conversion of glucosamine 6-PO4 to fructose 6-PO4 and ammonia. Arch Biochem Biophys 64:489–497
Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F (2010) The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75:1133–1144
Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H (2008) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464
Plumbridge J (1995) Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J 14:3958–3965
Reissig JL, Storminger JL, Leloir LF (1955) A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem 217:959–966
Rittmann D, Lindner SN, Wendisch VF (2008) Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol 74:6216–6222
Sambrook J, Russell D (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratoy Press, Cold Spring Harbor, NY
Schneider J, Wendisch VF (2010) Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 88:859–868
Schneider J, Niermann K, Wendisch VF (2011) Production of the amino acids l-glutamate, l-lysine, l-ornithine and l-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol 154:191–198
Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ (2006) Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124:381–391
Song Y, Matsumoto K, Tanaka T, Kondo A, Taguchi S (2013) Single-step production of polyhydroxybutyrate from starch by using alpha-amylase cell-surface displaying system of Corynebacterium glutamicum. J Biosci Bioeng 115:12–14
Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF (2005) Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol 71:5920–5928
Tateno T, Fukuda H, Kondo A (2007) Production of l-lysine from starch by Corynebacterium glutamicum displaying alpha-amylase on its cell surface. Appl Microbiol Biotechnol 74:1213–1220
Tateno T, Okada Y, Tsuchidate T, Tanaka T, Fukuda H, Kondo A (2009) Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl Microbiol Biotechnol 82:115–121
Tauch A, Kirchner O, Löffler B, Gotker S, Pühler A, Kalinowski J (2002) Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol 45:362–367
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:25–38
Toyoda K, Teramoto H, Inui M, Yukawa H (2008) Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl Microbiol Biotechnol 81:291–301
Typas A, Banzhaf M, Gross CA, Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136
Uehara T, Park JT (2004) The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J Bacteriol 186:7273–7279
Uhde A, Youn JW, Maeda T, Clermont L, Matano C, Krämer R, Wendisch VF, Seibold GM, Marin K (2013) Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl Microbiol Biotechnol 97:1679–1687
Wendisch VF (ed) (2007) Amino acid biosynthesis—pathways, regulation and metabolic engineering. Springer Verlag, Berlin
Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ (2000) Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J Bacteriol 182:3088–3096
Wendland J, Schaub Y, Walther A (2009) N-Acetylglucosamine utilization by Saccharomyces cerevisiae based on expression of Candida albicans NAG genes. Appl Environ Microbiol 75:5840–5845
White RJ (1968) Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J 106:847–858
White RJ (1970) The role of the phosphoenolpyruvate phosphotransferase system in the transport of N-acetyl-d-glucosamine by Escherichia coli. Biochem J 118:89–92
Wieschalka S, Blombach B, Eikmanns BJ (2012) Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol 94:449–459
Winnen B, Felce J, Saier MH Jr (2005) Genomic analyses of transporter proteins in Corynebacterium glutamicum and Corynebacterium efficiens. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL, pp 149–186
Yao W, Chu C, Deng X, Zhang Y, Liu M, Zheng P, Sun Z (2009) Display of alpha-amylase on the surface of Corynebacterium glutamicum cells by using NCgl1221 as the anchoring protein, and production of glutamate from starch. Arch Microbiol 191:751–759
Youn JW, Jolkver E, Krämer R, Marin K, Wendisch VF (2008) Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J Bacteriol 190:6458–6466
Youn JW, Jolkver E, Krämer R, Marin K, Wendisch VF (2009) Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J Bacteriol 191:5480–5488
Zahoor A, Lindner SN, Wendisch VF (2012) Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput Struct Biotechnol J 3:4
Acknowledgments
The authors thank Ute Meyer and Eva Glees (Institute of Biochemistry, University of Cologne, Germany) for excellent technical assistance, Jörn Kalinowski and Christian Rückert (Cebitec, University of Bielefeld, Germany) for granting early access to the C. glycinophilum draft genome sequence, and Jacqueline Plumbridge(Institut de Biologie Physico-Chimique, CNRS UPR9073, France) for providing the strain E. coli LR2-168 Work in the laboratories of the authors was funded in part by grants 0315589G and 0315589 F from BMBF in the CRP “Corynebacterium: improving flexibility and fitness for industrial production”.
Author information
Authors and Affiliations
Corresponding author
Additional information
C. Matano and A. Uhde contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 112 kb)
Rights and permissions
About this article
Cite this article
Matano, C., Uhde, A., Youn, JW. et al. Engineering of Corynebacterium glutamicum for growth and l-lysine and lycopene production from N-acetyl-glucosamine. Appl Microbiol Biotechnol 98, 5633–5643 (2014). https://doi.org/10.1007/s00253-014-5676-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5676-9