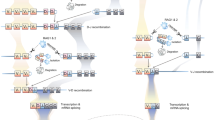
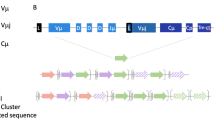
Abstract
Immunoglobulins and T cell receptors (TCR) have obvious structural similarities as well as similar immunogenetic diversification and selection mechanisms. Nevertheless, the two receptor systems and the loci that encode them are distinct in humans and classical murine models, and the gene segments comprising each repertoire are mutually exclusive. Additionally, while both B and T cells employ recombination-activating genes (RAG) for primary diversification, immunoglobulins are afforded a supplementary set of activation-induced cytidine deaminase (AID)-mediated diversification tools. As the oldest-emerging vertebrates sharing the same adaptive B and T cell receptor systems as humans, extant cartilaginous fishes allow a potential view of the ancestral immune system. In this review, we discuss breakthroughs we have made in studies of nurse shark (Ginglymostoma cirratum) T cell receptors demonstrating substantial integration of loci and diversification mechanisms in primordial B and T cell repertoires. We survey these findings in this shark model where they were first described, while noting corroborating examples in other vertebrate groups. We also consider other examples where the gnathostome common ancestry of the B and T cell receptor systems have allowed dovetailing of genomic elements and AID-based diversification approaches for the TCR. The cartilaginous fish seem to have retained this T/B cell plasticity to a greater extent than more derived vertebrate groups, but representatives in all vertebrate taxa except bony fish and placental mammals show such plasticity.



Similar content being viewed by others
References
Adams EJ, Chien YH, Garcia KC (2005) Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 308:227–231. https://doi.org/10.1126/science.1106885
Aghaallaei N, Bajoghli B (2018) Making thymus visible: understanding T-cell development from a new perspective. Front Immunol 9:375. https://doi.org/10.3389/fimmu.2018.00375
Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z (2005) Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310:1970–1973. https://doi.org/10.1126/science.1119420
Allison TJ, Garboczi DN (2002) Structure of gammadelta T cell receptors and their recognition of non-peptide antigens. Mol Immunol 38:1051–1061. https://doi.org/10.1016/S0161-5890(02)00034-2
Álvarez-Prado ÁF, Pérez-Durán P, Pérez-García A, Benguria A, Torroja C, de Yébenes VG, Ramiro AR (2018) A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J Exp Med 215:761–771. https://doi.org/10.1084/jem.20171738
Anderson MK, Shamblott MJ, Litman RT, Litman GW (1995) Generation of immunoglobulin light chain gene diversity in Raja erinacea is not associated with somatic rearrangement, an exception to a central paradigm of B cell immunity. J Exp Med 182:109–119. https://doi.org/10.1084/jem.182.1.109
Antonacci R, Vaccarelli G, Di Meo GP, Piccinni B, Miccoli MC, Cribiu EP, Perucatti A, Iannuzzi L, Ciccarese S (2007) Molecular in situ hybridization analysis of sheep and goat BAC clones identifies the transcriptional orientation of T cell receptor gamma genes on chromosome 4 in Bovids. Vet Res Commun 31:977–983. https://doi.org/10.1007/s11259-006-0202-x
Antonacci R, Mineccia M, Lefranc MP, Ashmaoui HM, Lanave C, Piccinni B, Pesole G, Hassanane MS, Massari S, Ciccarese S (2011) Expression and genomic analyses of Camelus dromedarius T cell receptor delta (TRD) genes reveal a variable domain repertoire enlargement due to CDR3 diversification and somatic mutation. Mol Immunol 48:1384–1396. https://doi.org/10.1016/j.molimm.2011.03.011
Arakawa H, Hauschild J, Buerstedde J-M (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301–1306. https://doi.org/10.1126/science.1067308
Augustin A, Sim G (1984) T-cell receptors generated via mutations are specific for various major histocompatibility antigens. Cell 39:5–12. https://doi.org/10.1016/0092-8674(84)90186-7
Bachl J, Wabl M (1995) Hypermutation in T cells questioned. Nature 375:285–286. https://doi.org/10.1038/375285c0
Bailey M, Christoforidou Z, Lewis M (2013) Evolution of immune systems: Specificity and autoreactivity. Autoimmun Rev 12(6):643–647. https://doi.org/10.1016/j.autrev.2012.10.007
Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109:S45–S55. https://doi.org/10.1016/S0092-8674(02)00675-X
Beetz S, Wesch D, Marischen L, Welte S, Oberg H-H, Kabelitz D (2008) Innate immune functions of human γδ T cells. Immunobiol 213:173–182. https://doi.org/10.1016/j.imbio.2007.10.006
Bilal S, Lie KK, Sæle Ø, Hordvik I (2018) T cell receptor alpha chain genes in the teleost ballan wrasse (Labrus bergylta) are subjected to somatic hypermutation. Front Immunol 9:1101. https://doi.org/10.3389/fimmu.2018.01101
Borgulya P, Kishi H, Uematsu Y, von Boehmer H (1992) Exclusion and inclusion of alpha and beta T cell receptor alleles. Cell 69:529–537. https://doi.org/10.1016/0092-8674(92)90453-j
Brady BL, Steinel NC, Bassing CH (2010) Antigen receptor allelic exclusion: an update and reappraisal. J Immunol 185:3801–3808. https://doi.org/10.4049/jimmunol.1001158
Brandes M, Willimann K, Bioley G, Lévy N, Eberl M, Luo M, Tampé R, Lévy F, Romero P, Moser B (2009) Cross-presenting human γδ T cells induce robust CD8+ αβ T cell responses. Proc Natl Acad Sci 106:2307–2312. https://doi.org/10.1073/pnas.0810059106
Brazeau MD, Friedman M (2015) The origin and early phylogenetic history of jawed vertebrates. Nature 520:490–497. https://doi.org/10.1038/nature14438
Breaux B, Hunter ME, Cruz-Schneider MP, Sena L, Bonde RK, Criscitiello MF (2018) The Florida manatee (Trichechus manatus latirostris) T cell receptor loci exhibit V subgroup synteny and chain-specific evolution. Dev Comp Immunol 85:71–85. https://doi.org/10.1016/j.dci.2018.04.007
Casetti R, Agrati C, Wallace M, Sacchi A, Martini F, Martino A, Rinaldi A, Malkovsky M (2009) Cutting edge: TGF-β1 and IL-15 induce FOXP3+ γδ regulatory T cells in the presence of antigen stimulation. J Immunol 183:3574–3577. https://doi.org/10.4049/jimmunol.0901334
Chang B, Casali P (1994) The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today 15:367–373. https://doi.org/10.1016/0167-5699(94)90175-9
Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF, Marchalonis JJ (2009) Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc Natl Acad Sci 106:8591–8596. https://doi.org/10.1073/pnas.0811283106
Chen H, Bernstein H, Ranganathan P, Schluter S (2012) Somatic hypermutation of TCR γ V genes in the sandbar shark. Dev Comp Immunol 37:176–183. https://doi.org/10.1016/j.dci.2011.08.018
Cheynier R, Henrichwark S, Wain Hobson S (1998) Somatic hypermutation of the T cell receptor V beta gene in microdissected splenic white pulps from HIV-1-positive patients. Eur J Immunol 28:1604–1610. https://doi.org/10.1002/(SICI)1521-4141(199805)28:05
Chien YH, Iwashima M, Kaplan KB, Elliot JF, Davis MM (1987) A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature 327:677–682. https://doi.org/10.1038/327677a0
Ciccarese S, Vaccarelli G, Lefranc MP, Tasco G, Consiglio A, Casadio R, Linguiti G, Antonacci R (2014) Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains. Dev Comp Immunol 46:300–313. https://doi.org/10.1016/j.dci.2014.05.001
Conticello SG, Thomas CJF, Petersen-Mahrt SK, Neuberger MS (2005) Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377. https://doi.org/10.1093/molbev/msi026
Conticello SG, Langlois MA, Yang Z, Neuberger MS (2007) DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94:37–73. https://doi.org/10.1016/S0065-2776(06)94002-4
Criscitiello M, Saltis M, Flajnik M (2006) An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci 103:5036–5041. https://doi.org/10.1073/pnas.0507074103
Criscitiello M, Flajnik M (2007) Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol 37:2683–2694. https://doi.org/10.1002/eji.200737263
Criscitiello MF, Wermenstam NE, Pilstrom L, McKinney EC (2004) Allelic polymorphism of T-cell receptor constant domains is widespread in fishes. Immunogenetics 55:818–824. https://doi.org/10.1007/s00251-004-0652-7
Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF (2010) Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol 184:6950–6960. https://doi.org/10.4049/jimmunol.0902774
Criscitiello MF, Ohta Y, Graham MD, Eubanks JO, Chen PL, Flajnik MF (2012) Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev Comp Immunol 36:521–533. https://doi.org/10.1016/j.dci.2011.09.008
Criscitiello MF, Dickman MB, Samuel JE, de Figueiredo P (2013) Tripping on acid: trans-kingdom perspectives on biological acids in immunity and pathogenesis. PLOS Pathog 9:e1003402. https://doi.org/10.1371/journal.ppat.1003402
Criscitiello MF (2014) Shark T cell receptors. In: Smith SL, Sim RB, Flajnik MF (eds) Immunobiology of the Shark, 1st edn. CRC Press, Boca Raton, USA. doi: https://doi.org/10.1201/b17773
Das S, Li J, Hirano M, Sutoh Y, Herrin BR, Cooper MD (2015) Evolution of two prototypic T cell lineages. Cellular immunol 296:87–94. https://doi.org/10.1016/j.cellimm.2015.04.007
Deiss TC, Vadnais M, Wang F, Chen PL, Torkamani A, Mwangi W, Lefranc MP, Criscitiello MF, Smider VV (2017) Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol Immunol. https://doi.org/10.1038/cmi.2017.117
Deiss TC, Breaux B, Ott JA, Daniel RA, Chen PL, Castro CD, Ohta Y, Flajnik MF, Criscitiello MF (2019) Ancient use of Ig variable domains contributes significantly to the TCRδ repertoire. J Immunol 203:1265–1275. https://doi.org/10.4049/jimmunol.1900369
Deng L, Velikovsky CA, Xu G, Iyer L, Tasumi S, Kerzic M, Flajnik M, Aravind L, Pancer Z, Mariuzza R (2010) A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci 107:13408–13413. https://doi.org/10.1073/pnas.1005475107
Diaz M, Flajnik MF (1998) Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol Rev 162:13–24. https://doi.org/10.1111/j.1600-065x.1998.tb01425.x
Diaz M, Greenberg A, Flajnik M (1998) Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci 95:14343–14348. https://doi.org/10.1073/pnas.95.24.14343
Diaz M, Velez J, Singh M, Cerny J, Flajnik MF (1999) Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol 11:825–833. https://doi.org/10.1093/intimm/11.5.825
Diaz M, Flajnik MF, Klinman N (2001) Evolution and the molecular basis of somatic hypermutation of antigen receptor genes. Phil Trans R Soc B Biol Sci 356:67–72
Dooley H, Flajnik MF (2005) Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol 35:936–945. https://doi.org/10.1002/eji.200425760
Dooley H, Flajnik MF (2006) Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 30:43–56. https://doi.org/10.1016/j.dci.2005.06.022
Dooley H, Stanfield RL, Brady RA, Flajnik MF (2006a) First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci 103:1846–1851. https://doi.org/10.1073/pnas.0508341103
Dooley H, Stanfield RL, Brady RA, Flajnik MF (2006b) First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci USA 103:1846–1851
Fahl SP, Coffey F, Wiest DL (2014) Origins of γδ T cell effector subsets: a riddle wrapped in an enigma. J Immunol 193:4289–4294. https://doi.org/10.4049/jimmunol.1401813
Flajnik MF, Rumfelt LL (2000) The immune system of cartilaginous fish. Curr Top Microbiol Immunol 248:249–270. https://doi.org/10.1007/978-3-642-59674-2_11
Flajnik MF (2002) Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2:688–698. https://doi.org/10.1038/nri889
Flajnik MF (2014) Re-evaluation of the Immunological Big Bang. Curr Biol 24:R1060–R1065. https://doi.org/10.1016/j.cub.2014.09.070
Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:438–453. https://doi.org/10.1038/s41577-018-0003-9
Gascoigne NRJ, Alam SM (1999) Allelic exclusion of the T cell receptor α-chain: developmental regulation of a post-translational event. Semin Immunol 11:337–347. https://doi.org/10.1006/smim.1999.0190
Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71:101–132. https://doi.org/10.1146/annurev.biochem.71.090501.150203
Germain RN (2002) T-cell development and the CD4–CD8 lineage decision. Nat Rev Immunol 2:309–322. https://doi.org/10.1038/nri798
Gober H-J, Kistowska M, Angman L, Jenö P, Mori L, De Libero G (2003) Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med 197:163–168. https://doi.org/10.1084/jem.20021500
Good RA, Finstad J (1966) The Phylogenetic Development of Immune Responses and the Germinal Center System. In: Cottier H, Odartchenko N, Schindler R, Congdon CC (eds) Germinal Centers in Immune Responses. Springer-Verlag, New York Inc, University of Bern, Switzerland
Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF (1995) A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374:168–173. https://doi.org/10.1038/374168a0
Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD (2009) Dual nature of the adaptive immune system in lampreys. Nature 459:796–801. https://doi.org/10.1038/nature08068
Hackett J Jr, Stebbins C, Rogerson B, Davis MM, Storb U (1992) Analysis of a T cell receptor gene as a target of the somatic hypermutation mechanism. J Exp Med 176:225–231. https://doi.org/10.1084/jem.176.1.225
Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD, Tatsumi K, Tanaka K, Motone F, Kageyama Y, Nozu R, Adachi N, Nishimura O, Nakagawa R, Tanegashima C, Kiyatake I, Matsumoto R, Murakumo K, Nishida K, Terakita A, Kuratani S, Sato K, Hyodo S, Kuraku S (2018) Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat Ecol Evol 2:1761–1771. https://doi.org/10.1038/s41559-018-0673-5
Hayday AC, Vantourout P (2020) The innate biologies of adaptive antigen receptors. Ann Rev Immunol 38:487–510. https://doi.org/10.1146/annurev-immunol-102819-023144
Hsu E, Criscitiello MF (2006) Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol 177:2452–2462. https://doi.org/10.4049/jimmunol.177.4.2452
Hsu E (2009) V(D)J Recombination: Of Mice and Sharks. Adv Exp Med Biol 650:166–179. https://doi.org/10.1007/978-1-4419-0296-2_14
Hsu E (2018) Immune system receptors in vertebrates: immunoglobulins. Reference Module in Life Sciences. https://doi.org/10.1016/B978-0-12-809633-8.20721-8
Huesmann M, Scott B, Kisielow P, von Boehmer H (1991) Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell 66:533–540. https://doi.org/10.1016/0092-8674(81)90016-7
Iacoangeli A, Lui A, Haines A, Ohta Y, Flajnik M, Hsu E (2017) Evidence for Ig light chain isotype exclusion in shark B lymphocytes suggests ordered mechanisms. J Immunol 199:1875–1885. https://doi.org/10.4049/jimmunol.1700762
Jack R, Du Pasquier L (2019) The Triumph of Individualism: Evolution of Somatically Generated Adaptive Immune Systems. Evolutionary Concepts in Immunology. Springer International Publishing, Cham. doi: https://doi.org/10.1007/978-3-030-18667-8_4
Janvier P (2011) Comparative anatomy: all vertebrates do have vertebrae. Curr Biol 21:R661–R663. https://doi.org/10.1016/j.cub.2011.07.014
Jhunjhunwala S, van Zelm MC, Peak MM, Murre C (2009) Chromatin architecture and the generation of antigen receptor diversity. Cell 138:435–448. https://doi.org/10.1016/j.cell.2009.07.016
Kabelitz D (2011) γδ T-cells: cross-talk between innate and adaptive immunity. Cell Mol Life Sci 68:2331. https://doi.org/10.1007/s00018-011-0696-4
Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M (2010) Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci 107:14304–14308. https://doi.org/10.1073/pnas.1001910107
Kazen AR, Adams EJ (2011) Evolution of the V, D, and J gene segments used in the primate γδ T-cell receptor reveals a dichotomy of conservation and diversity. Proc Natl Acad Sci 108:E332–E340. https://doi.org/10.1073/pnas.1105105108
Kikutani H, Inui S, Sato R, Barsumian EL, Owaki H, Yamasaki K, Kaisho T, Uchibayashi N, Hardy RR, Hirano T, Tsunasawa S, Sakiyama F, Suemura M, Kishimoto T (1986) Molecular structure of human lymphocyte receptor for immunoglobulin E. Cell 47:657–665. https://doi.org/10.1016/0092-8674(86)90508-8
Kondo K, Ohigashi I, Takahama Y (2019) Thymus machinery for T-cell selection. Int Immunol 31:119–125. https://doi.org/10.1093/intimm/dxy081
Kreslavsky T, Gleimer M, Garbe AI, von Boehmer H (2010) αβ versus γδ fate choice: counting the T-cell lineages at the branch point. Immunol Rev 238:169–181. https://doi.org/10.1111/j.1600-065X.2010.00947.x
Kronenberg M, Siu G, Hood LE, Shastri N (1986) The molecular genetics of the T cell antigen receptor and T cell antigen recognition. Ann Rev Immunol 4:529–591. https://doi.org/10.1146/annurev.iy.04.040186.002525
Kuklina EM (2006) Revision of the antigen receptor of T-lymphocytes. Biochemistry 71:827–837. https://doi.org/10.1134/S0006297906080025
Kuo TC, Schlissel MS (2009) Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol 21:173–178. https://doi.org/10.1016/j.coi.2009.03.008
Lafaille JJ, Haas W, Coutinho A, Tonegawa S (1990) Positive selection of γδ T cells. Immunol Today 11:75–78. https://doi.org/10.1016/0167-5699(90)90030-D
Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E (2002) Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16:571–582. https://doi.org/10.1016/s1074-7613(02)00300-x
Lefranc M-P (2014) Immunoglobulin and T cell receptor genes: IMGT and the birth and rise of immunoinformatics. Front Immunol 5:22–22. https://doi.org/10.3389/fimmu.2014.00022
Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 27:55–77. https://doi.org/10.1016/S0145-305X(02)00039-3
Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD (2004) The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev 18:1–11. https://doi.org/10.1101/gad.1161904
Liu M-C, Liao W-Y, Buckley KM, Yang SY, Rast JP, Fugmann SD (2018) AID/APOBEC-like cytidine deaminases are ancient innate immune mediators in invertebrates. Nat Commun 9:1948–1948. https://doi.org/10.1038/s41467-018-04273-x
Livak F, Schatz DG (1996) T-cell receptor alpha locus V(D)J recombination by-products are abundant in thymocytes and mature T cells. Mol Cell Biol 16:609–618. https://doi.org/10.1128/mcb.16.2.609
Luer C, Walsh CJ, Bodine AB, Wyffels JT, Scott TR (1995) The elasmobranch thymus: anatomical, histological, and preliminary functional characterization. J Exp Zool 273:342–354. https://doi.org/10.1002/jez.1402730408
MacLennan ICM (1994) Germinal Centers Ann Rev Immunol 12:117–139. https://doi.org/10.1146/annurev.iy.12.040194.001001
Magor BG (2015) Antibody affinity maturation in fishes-our current understanding. Biology 4:512–524. https://doi.org/10.3390/biology4030512
Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E (2005) Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J Immunol 175:8105–8115. https://doi.org/10.4049/jimmunol.175.12.8105
Marra NJ, Stanhope MJ, Jue NK, Wang M, Sun Q, Pavinski Bitar P, Richards VP, Komissarov A, Rayko M, Kliver S, Stanhope BJ, Winkler C, O’Brien SJ, Antunes A, Jorgensen S, Shivji MS (2019) White shark genome reveals ancient elasmobranch adaptations associated with wound healing and the maintenance of genome stability. Proc Natl Acad Sci 116:4446–4455. https://doi.org/10.1073/pnas.1819778116
Martin AP (1999) Substitution rates of organelle and nuclear genes in sharks: implicating metabolic rate (again). Mol Cell Biol 16:996–1002. https://doi.org/10.1093/oxfordjournals.molbev.a026189
Maul RW, Gearhart PJ (2010) Chapter six - AID and Somatic Hypermutation. In Alt FW (ed.) Adv Immunol. Academic Pressdoi: https://doi.org/10.1016/S0065-2776(10)05006-6
McGargill MA, Derbinski JM, Hogquist KA (2000) Receptor editing in developing T cells. Nat Immunol 1:336–341. https://doi.org/10.1038/79790
Morisawa T, Marusawa H, Ueda Y, Iwai A, Okazaki I-m, Honjo T, Chiba T (2008) Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. Int J Cancer 123:2735–2740. https://doi.org/10.1002/ijc.23853
Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563. https://doi.org/10.1016/S0092-8674(00)00078-7
Murphy K, Weaver C (2017) Janeway’s Immunobiology, 9th edn. Garland Science, New York
Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, Kanagawa O, Takahama Y (2012) Thymic nurse cells provide microenvironment for secondary T cell receptor α rearrangement in cortical thymocytes. Proc Natl Acad Sci 109:20572–20577. https://doi.org/10.1073/pnas.1213069109
Neely HR, Guo J, Flowers EM, Criscitiello MF, Flajnik MF (2018) “Double-duty” conventional dendritic cells in the amphibian Xenopus as the prototype for antigen presentation to B cells. Eur J Immunol 48:430–440. https://doi.org/10.1002/eji.201747260
Nielsen J, Hedeholm RB, Heinemeier J, Bushnell PG, Christiansen JS, Olsen J, Ramsey CB, Brill RW, Simon M, Steffensen KF, Steffensen JF (2016) Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 353:702–704. https://doi.org/10.1126/science.aaf1703
Odegard VH, Schatz DG (2006) Targeting of somatic hypermutation. Nat Rev Immunol 6:573–583. https://doi.org/10.1038/nri1896
Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF (2002) Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J Immunol 168:771–781. https://doi.org/10.4049/jimmunol.168.2.771
Ohta Y, Flajnik M (2006) IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci 103:10723–10728. https://doi.org/10.1073/pnas.0601407103
Ohta Y, Kasahara M, O’Connor TD, Flajnik MF (2019) Inferring the “Primordial Immune Complex”: origins of MHC class I and antigen receptors revealed by comparative genomics. J Immunol 203:1882–1896. https://doi.org/10.4049/jimmunol.1900597
Okazaki I-m, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T (2003) Constitutive expression of AID leads to tumorigenesis. J Exp Med 197:1173–1181. https://doi.org/10.1084/jem.20030275
Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, Criscitiello MF (2018) Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. eLife 7:e28477. doi: https://doi.org/10.7554/eLife.28477
Ott JA, Harrison J, Flajnik MF, Criscitiello MF (2020) Nurse shark T cell receptors employ somatic hypermutation preferentially to alter alpha/delta variable segments associated with alpha constant region. Eur J Immunol 50:1307–1320. https://doi.org/10.1002/eji.201948495
Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD (2005) Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci 102:9224–9229. https://doi.org/10.1073/pnas.0503792102
Parra ZE, Baker ML, Schwarz R, Deakin J, Lindblad-Toh K, Miller RD (2007) A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci 104(23):9776–9781. https://doi.org/10.1073/pnas.0609106104
Parra ZE, Baker ML, Hathaway J, Lopez AM, Trujillo J, Sharp A, Miller RD (2008) Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics 9:111. https://doi.org/10.1186/1471-2164-9-111
Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD (2010) The dynamic TCRdelta: TCRdelta chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur J Immunol 40:2319–2329. https://doi.org/10.1002/eji.201040515
Parra ZE, Lillie M, Miller RD (2012) A model for the evolution of the mammalian t-cell receptor α/δ and μ loci based on evidence from the duckbill Platypus. Mol Biol Evol 29:3205–3214. https://doi.org/10.1093/molbev/mss128
Parra ZE, Miller RD (2012) Comparative analysis of the chicken TCRα/δ locus. Immunogenetics 64:641–645. https://doi.org/10.1007/s00251-012-0621-5
Parra ZE, Mitchell K, Dalloul RA, Miller RD (2012) A second TCRdelta locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRdelta and TCRmu genes in tetrapods. J Immunol 188:3912–3919. https://doi.org/10.4049/jimmunol.1103521
Qin H, Suzuki K, Nakata M, Chikuma S, Izumi N, Thi Huong L, Maruya M, Fagarasan S, Busslinger M, Honjo T, Nagaoka H (2011) Activation-induced cytidine deaminase expression in CD4+ T cells is associated with a unique IL-10-producing subset that increases with age. PLoS ONE 6:e29141. https://doi.org/10.1371/journal.pone.0029141
Qin T, Zhao H, Zhu H, Wang D, Du W, Hao H (2015) Immunoglobulin genomics in the prairie vole (Microtus ochrogaster). Immunol Lett 166:79–86. https://doi.org/10.1016/j.imlet.2015.06.001
Rios FM, Zimmerman LM (2015) Immunology of Reptiles. John Wiley & Sons, Ltd
Rocco L, Morescalchi MA, Costagliola D, Stingo V (2002) Karyotype and genome characterization in four cartilaginous fishes. Gene 295:289–298. https://doi.org/10.1016/s0378-1119(02)00730-8
Rocco L, Liguori I, Costagliola D, Morescalchi MA, Tinti F, Stingo V (2007) Molecular and karyological aspects of Batoidea (Chondrichthyes, Elasmobranchi) phylogeny. Gene 389:80–86. https://doi.org/10.1016/j.gene.2006.09.024
Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z (2007) Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol 8:647–656. https://doi.org/10.1038/ni1463
Rucci F, Cattaneo L, Marrella V, Sacco MG, Sobacchi C, Lucchini F, Nicola S, Bella SD, Villa ML, Imberti L, Gentili F, Montagna C, Tiveron C, Tatangelo L, Facchetti F, Vezzoni P, Villa A (2006) Tissue-specific sensitivity to AID expression in transgenic mouse models. Gene 377:150–158. https://doi.org/10.1016/j.gene.2006.03.024
Rumfelt L, McKinney E, Taylor E, Flajnik M (2002) The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol 56:130–148. https://doi.org/10.1046/j.1365-3083.2002.01116.x
Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F, Canapa A, Cheng J-F, Amemiya CT (2014) Genome complexity in the coelacanth is reflected in its adaptive immune system. J Exp Zool Part B 322:438–463. https://doi.org/10.1002/jez.b.22558
Saini J, Hershberg U (2015) B cell variable genes have evolved their codon usage to focus the targeted patterns of somatic mutation on the complementarity determining regions. Mol Immunol 65:157–167. https://doi.org/10.1016/j.molimm.2015.01.001
Schatz DG (2004) V(D)J recombination. Immunol Rev 200:5–11. https://doi.org/10.1111/j.0105-2896.2004.00173.x
Stingo V, Rocco L (2001) Selachian cytogenetics: a review. Genetica 111:329–347. https://doi.org/10.1023/a:1013747215866
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302:575–581. https://doi.org/10.1038/302575a0
Vaccarelli G, Miccoli MC, Antonacci R, Pesole G, Ciccarese S (2008) Genomic organization and recombinational unit duplication-driven evolution of ovine and bovine T cell receptor gamma loci. BMC Genomics 9:81. https://doi.org/10.1186/1471-2164-9-81
Vaccarelli G, Antonacci R, Tasco G, Yang F, Giordano L, El Ashmaoui HM, Hassanane MS, Massari S, Casadio R, Ciccarese S (2012) Generation of diversity by somatic mutation in the Camelus dromedarius T-cell receptor gamma variable domains. Eur J Immunol 42:3416–3428. https://doi.org/10.1002/eji.201142176
Venkatesh B, Kirkness EF, Loh Y-H, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S (2007) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLOS Biol 5:e101. https://doi.org/10.1371/journal.pbio.0050101
Venkatesh B, Lee AP, Ravi V (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179. https://doi.org/10.1038/nature12826
Vernooij BTM, Lenstra JA, Wang K, Hood L (1993) Organization of the murine T-cell receptor γ locus. Genomics 17:566–574. https://doi.org/10.1006/geno.1993.1373
Vitetta E, Berton M, Burger C, Kepron M, Wa L, Yin X (1991) Memory B and T cells. Ann Rev Immunol 9:193–217. https://doi.org/10.1146/annurev.iy.09.040191.001205
Wang X, Parra ZE, Miller RD (2011) Platypus TCRmu provides insight into the origins and evolution of a uniquely mammalian TCR locus. J Immunol 187:5246–5254. https://doi.org/10.4049/jimmunol.1101113
Wang X, Huang J, Wang P, Wang R, Wang C, Yu D, Ke C, Huang T, Song Y, Bai J, Li K, Ren L, Miller RD, Han H, Zhou X, Zhao Y (2020) Analysis of the Chinese alligator TCRalpha/delta loci reveals the evolutionary pattern of atypical TCRdelta/TCRmu in tetrapods. J Immunol 205:637–647. https://doi.org/10.4049/jimmunol.2000257
Zapata A, Leceta J, Barrutia MG (1981) Ultrastructure of splenic white pulp of the turtle, Mauremys caspica. Cell Tissue Res 220:845–855. https://doi.org/10.1007/bf00210466
Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, Schatz DG (2019) Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569:79–84. https://doi.org/10.1038/s41586-019-1093-7
Zheng B, Xue W, Kelsoe G (1994) Locus-specific somatic hypermutation in germinal centre T cells. Nature 372:556–559. https://doi.org/10.1038/372556a0
Zhu C, Hsu E (2010) Error-prone DNA repair activity during somatic hypermutation in shark B lymphocytes. J Immunol 185:5336–5347. https://doi.org/10.4049/jimmunol.1000779
Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E (2012) Origin of immunoglobulin isotype switching. Curr Biol. https://doi.org/10.1016/j.cub.2012.03.060
Zimmerman LM, Vogel LA, Bowden RM (2010) Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol 213:661–671. https://doi.org/10.1242/jeb.038315
Funding
This work was supported by grants from the NIH to MFC (AI56963) and MFF (AI027877 and AI140326) and the NSF to MFC (IOS-1257829 and IOS-1656870).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of Topical Collection on Fish Immunology
Rights and permissions
About this article
Cite this article
Ott, J.A., Ohta, Y., Flajnik, M.F. et al. Lost structural and functional inter-relationships between Ig and TCR loci in mammals revealed in sharks. Immunogenetics 73, 17–33 (2021). https://doi.org/10.1007/s00251-020-01183-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-020-01183-5