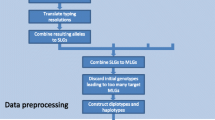
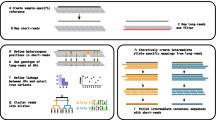
Abstract
Regardless of sampling depth, accurate genotype imputation is limited in regions of high polymorphism which often have a heavy-tailed haplotype frequency distribution. Many rare haplotypes are thus unobserved. Statistical methods to improve imputation by extending reference haplotype distributions using linkage disequilibrium patterns that relate allele and haplotype frequencies have not yet been explored. In the field of unrelated stem cell transplantation, imputation of highly polymorphic human leukocyte antigen (HLA) genes has an important application in identifying the best-matched stem cell donor when searching large registries totaling over 28,000,000 donors worldwide. Despite these large registry sizes, a significant proportion of searched patients present novel HLA haplotypes. Supporting this observation, HLA population genetic models have indicated that many extant HLA haplotypes remain unobserved. The absent haplotypes are a significant cause of error in haplotype matching. We have applied a Bayesian inference methodology for extending haplotype frequency distributions, using a model where new haplotypes are created by recombination of observed alleles. Applications of this joint probability model offer significant improvement in frequency distribution estimates over the best existing alternative methods, as we illustrate using five-locus HLA frequency data from the National Marrow Donor Program registry. Transplant matching algorithms and disease association studies involving phasing and imputation of rare variants may benefit from this statistical inference framework.





Similar content being viewed by others
References
Anasetti C (2012) The ever elusive permissive mismatch. Biol Blood Marrow Transplant 18:657–658
Browning SR, Weir BS (2010) Population structure with localized haplotype clusters. Genetics 185:1337–1344
Chi EC, Zhou H, Chen GK, Del Vecchyo DO, Lange K (2013) Genotype imputation via matrix completion. Genome Res 23:509–518
Consortium IH (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58
Dehn J, Arora M, Spellman S, Setterholm M, Horowitz M, Confer D, Weisdorf D (2008) Unrelated donor hematopoietic cell transplantation: factors associated with a better HLA match. Biol Blood Marrow Transplant 14(12):1334–1340. https://doi.org/10.1016/j.bbmt.2008.09.009
Dehn J, Setterholm M, Buck K, Kempenich J, Beduhn B, Gragert L, Madbouly A, Fingerson S, Maiers M (2016) HapLogic: a predictive human leukocyte antigen-matching algorithm to enhance rapid identification of the optimal unrelated hematopoietic stem cell sources for transplantation. Biol Blood Marrow Transplant 22(11):2038–2046
Dilthey A, Cox C, Iqbal Z, Nelson MR, McVean G (2015) Improved genome inference in the MHC using a population reference graph. Nat Genet 47:682–688
du Preez JA (1998) Efficient training of high-order hidden Markov models using first-order representations. Comput Speech Lang 12:23–39
Eberhard HP, Feldmann U, Bochtler W, Baier D, Rutt C, Schmidt AH, Müller CR (2010) Estimating unbiased haplotype frequencies from stem cell donor samples typed at heterogeneous resolutions: a practical study based on over 1 million German donors. Tissue Antigens 76:352–361
Eberhard HP, Madbouly A, Gourraud P, Balère M, Feldmann U, Gragert L, Maldonado Torres H, Pingel J, Schmidt A, Steiner D (2013) Comparative validation of computer programs for haplotype frequency estimation from donor registry data. Tissue Antigens 82:93–105
Erlich H (2012) HLA DNA typing: past, present, and future. Tissue Antigens 80:1–11
Ewens W (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3(1):87–112
Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927
Gragert L, Madbouly A, Freeman J, Maiers M (2013) Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 74:1313–1320
Hansen JA, Yamamoto K, Petersdorf E, Sasazuki T (1999) The role of {HLA} matching in hematopoietic cell transplantation. Rev Immunogenet 1:359–373
Hastie T, Tibshirani R, Friedman J (2009) Unsupervised learning. Springer, The elements of statistical learning
Hawley ME, Kidd KK (1995) {HAPLO:} a program using the {EM} algorithm to estimate the frequencies of multi-site haplotypes. J Hered 86:409–411
Hellinger E (1909) Neue Begr{ü}ndung der Theorie quadratischer Formen von unendlichvielen Ver{ä}nderlichen. Journal f{ü}r die reine und angewandte Mathematik 136:210–271
Hoerl AE, Kennard RW (1970) Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12:55–67
Hou L, Vierra-Green C, Lazaro A, Brady C, Haagenson M, Spellman S, Hurley C (2017) Limited HLA sequence variation outside of antigen recognition domain exons of 360 10 of 10 matched unrelated hematopoietic stem cell transplant donor-recipient pairs. HLA 89:39–46
Klitz W, Hedrick P, Louis EJ (2012) New reservoirs of HLA alleles: pools of rare variants enhance immune defense. Trends Genet 28:480–486
Kollman C, Maiers M, Gragert L, Müller C, Setterholm M, Oudshoorn M, Hurley CK (2007) Estimation of {HLA-A}, -B, -{DRB1} haplotype frequencies using mixed resolution data from a National Registry with selective retyping of volunteers. Hum Immunol 68:950–958
Kulkarni S, Martin MP, Carrington M (2008) The Yin and Yang of HLA and KIR in human disease. Elsevier, Seminars in immunology
Kullback S, Leibler RA (1951) On information and sufficiency. Ann Math Stat 22:79–86
Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, Fernandez-Vina M, Flomenberg N, Horowitz M, Hurley CK, Noreen H, Oudshoorn M, Petersdorf E, Setterholm M, Spellman S, Weisdorf D, Williams TM, Anasetti C (2007) High-resolution donor-recipient {HLA} matching contributes to the success of unrelated donor marrow transplantation. Blood 110:4576–4583
Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF (2013) Common and well-documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens 81:194–203
Mack SJ, Tu B, Lazaro A, Yang R, Lancaster AK, Cao K, Ng J, Hurley CK (2009) {HLA-A}, -B, -C, and -{DRB1} allele and haplotype frequencies distinguish eastern European Americans from the general European American population. Tissue Antigens 73:17–32
Maiers M, Gragert L, Klitz W (2007) High-resolution {HLA} alleles and haplotypes in the United States population. Hum Immunol 68:779–788
Maiers M, Gragert L, Madbouly A, Steiner D, Marsh SGE, Gourraud P-A, Oudshoorn M, van der Zanden H, Schmidt AH, Pingel J, Hofmann J, Müller C, Eberhard H-P (2013) 16(th) {IHIW:} global analysis of registry {HLA} haplotypes from 20 million individuals: report from the {IHIW} registry diversity group. Int J Immunogenet 40:66–71
Maiers M, Hurley C, Perlee L, Fernandez-Vina M, Baisch J, Cook D, Fraser P, Heine U, Hsu S, Leffell M (1999) Maintaining updated DNA-based HLA assignments in the National Marrow Donor Program Bone Marrow Registry. Rev Immunogenet 2:449–460
Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11:499–511
Martin AM, Freitas EM, Witt CS, Christiansen FT (2000) The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics 51:268–280
Niu T (2004) Algorithms for inferring haplotypes. Genet Epidemiol 27:334–347
Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, Jayaraman J, Wroblewski EE, Trowsdale J, Rajalingam R (2016) Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet 99:375–391
Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A (2009) A comprehensive evaluation of {SNP} genotype imputation. Hum Genet 125:163–171
Paunić V, Gragert L, Schneider J, Mueller C, Maiers M (2016) Charting improvements in US registry HLA typing ambiguity using a typing resolution score. Hum Immunol 77:542–549
Petersdorf EW, Anasetti C, Martin PJ, Gooley T, Radich J, Malkki M, Woolfrey A, Smith A, Mickelson E, Hansen JA (2004) Limits of {HLA} mismatching in unrelated hematopoietic cell transplantation. Blood 104:2976–2980
Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z (2007) MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med 4:e8
Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG (2014) The IPD and IMGT/HLA database: allele variant databases. Nucleic acids research:gku1161
Schafer JL, Graham JW (2002) Missing data: our view of the state of the art. Psychol Methods 7:147–177
Siva N (2008) 1000 Genomes project. Nat Biotechnol 26:256–256
Slater N, Louzoun Y, Gragert L, Maiers M, Chatterjee A, Albrecht M (2015) Power laws for heavy-tailed distributions: modeling allele and haplotype diversity for the National Marrow Donor Program. PLoS Comput Biol 11(4)
Spellman SR, Eapen M, Logan BR, Mueller C, Rubinstein P, Setterholm MI, Woolfrey AE, Horowitz MM, Confer DL, Hurley CK (2012) A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood 120:259–265
Templeton AR, Sing CF (1993) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 134:659–669
Trowsdale J, Knight JC (2013) Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet 14:301–323
Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E, Lebedeva T, Yu N, Stewart M, Noreen H (2012) Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS One 7:e47491
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Louzoun, Y., Alter, I., Gragert, L. et al. Modeling coverage gaps in haplotype frequencies via Bayesian inference to improve stem cell donor selection. Immunogenetics 70, 279–292 (2018). https://doi.org/10.1007/s00251-017-1040-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1040-4