Abstract
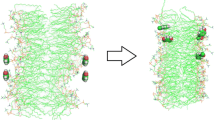
In this work, the effects of the anti-hypertensive drug amlodipine in native and PEGylated forms on the malfunctioning of negatively charged lipid bilayer cell membranes constructed from DMPS or DMPS + DMPC were studied by molecular dynamics simulation. The obtained results indicate that amlodipine alone aggregates and as a result its diffusion into the membrane is retarded. In addition, due to their large size aggregates of the drug can damage the cell, rupturing the cell membrane. It is shown that PEGylation of amlodipine prevents this aggregation and facilitates its diffusion into the lipid membrane. The interaction of the drug with negatively charged membranes in the presence of an aqueous solution of NaCl, as the medium, is investigated and its effects on the membrane are considered by evaluating the structural properties of the membrane such as area per lipid, thickness, lipid chain order and electrostatic potential difference between bulk solution and lipid bilayer surface. The effect of these parameters on the diffusion of the drug into the cell is critically examined and discussed.








Similar content being viewed by others
References
Alsop RJ et al (2016) The lipid bilayer provides a site for cortisone crystallization at high cortisone concentrations. Sci Rep 6:22425
Amjad-Iranagh S et al (2013) Effects of protein binding on a lipid bilayer containing local anesthetic articaine, and the potential of mean force calculation: a molecular dynamics simulation approach. J Mol Model 19:3831–3842
Bunker A (2012) Poly (ethylene glycol) in drug delivery, why does it work, and can we do better? All atom molecular dynamics simulation provides some answers. Phys Proc 34:24–33
Caron G et al (2004) Ionization, lipophilicity, and molecular modeling to investigate permeability and other biological properties of amlodipine. Bioorg Med Chem 12:6107–6118
Carrillo J-MY, Dobrynin AV (2011) Layer-by-layer assembly of charged nanoparticles on porous substrates: molecular dynamics simulations. ACS Nano 5:3010–3019
Carrozzino JM, Khaledi MG (2004) Interaction of basic drugs with lipid bilayers using liposome electrokinetic chromatography. Pharm Res 21:2327–2335
Lopez Cascales J, de la Torre JG (1997) Effect of lithium and sodium ions on a charged membrane of dipalmitoylphosphatidylserine: a study by molecular dynamics simulation. Biochim Biophys Acta (BBA) Biomembr 1330:145–156
Chen X-J, Liang Q (2017) Combined effects of headgroup charge and tail unsaturation of lipids on lateral organization and diffusion of lipids in model biomembranes. Chin Phys B 26:048701
Cherstvy A (2007) Electrostatics of DNA complexes with cationic lipid membranes. J Phys Chem B 111:7914–7927
Cordomi A, Edholm O, Perez JJ (2008) Effect of ions on a dipalmitoyl phosphatidylcholine bilayer. A molecular dynamics simulation study. J Phys Chem B 112:1397–1408
Das A, Adhikari C, Chakraborty A (2017) Interaction of different divalent metal ions with lipid bilayer: impact on the encapsulation of doxorubicin by lipid bilayer and lipoplex mediated deintercalation. J Phys Chem B 121:1854–1865
Davis CH, Berkowitz ML (2009) Structure of the Amyloid-β (1–42) monomer absorbed to model phospholipid bilayers: a molecular dynamics study. J Phys Chem B 113:14480–14486
Ergun S et al (2014) Agomelatine strongly interacts with zwitterionic DPPC and charged DPPG membranes. Biochim Biophys Acta (BBA) Biomembr 1838:2798–2806
García Daza FA, Mackie AD (2017) Coarse-grained simulations of modified Jeffamine ED900 micelles. Molecular Simulation 44:1–8
Gotrane DM et al (2010) A novel method for resolution of amlodipine. Org Process Res Dev 14:640–643
Gurtovenko AA, Vattulainen I (2008) Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. J Phys Chem B 112:1953–1962
Gurtovenko AA et al (2005) Effect of monovalent salt on cationic lipid membranes as revealed by molecular dynamics simulations. J Phys Chem B 109:21126–21134
Hess B et al (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hess B et al (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Hoda N et al (2016) Curcumin specifically binds to the human calcium–calmodulin-dependent protein kinase IV: fluorescence and molecular dynamics simulation studies. J Biomol Struct Dyn 34:572–584
Hyvonen M (2003) Molecular dynamics simulations on phospholipid membranes, PhD Thesis, Oulun Yliopisto Institute, University of Oulu, Finland
Ionov M et al (2011) Interaction of cationic phosphorus dendrimers (CPD) with charged and neutral lipid membranes. Coll Surf B 82:8–12
Jalili S, Saeedi M (2017) Study of procaine and tetracaine in the lipid bilayer using molecular dynamics simulation. Eur Biophys J 46:265–282
Jalili S et al (2015) Free energy simulations of amylin I26P mutation in a lipid bilayer. Eur Biophys J 44:37–47
Jean-François F et al (2008) Aggregation of cateslytin β-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry 47:6394–6402
Jørgensen AM et al (2007) Molecular dynamics simulations of Na+/Cl−-dependent neurotransmitter transporters in a membrane-aqueous system. ChemMedChem 2:827–840
Komura S, Shirotori H, Kato T (2003) Phase behavior of charged lipid bilayer membranes with added electrolyte. J Chem Phys 119:1157–1164
Lee BL, Kuczera K (2017) Simulating the free energy of passive membrane permeation for small molecules. Mol Simul. https://doi.org/10.1080/08927022.2017.1407029
Liu H et al (2016) Probing the structure and dynamics of caveolin-1 in a caveolae-mimicking asymmetric lipid bilayer model. Eur Biophys J 45:511–521
Lolicato F et al (2015) Resveratrol interferes with the aggregation of membrane-bound human-IAPP: a molecular dynamics study. Eur J Med Chem 92:876–881
Lopez Cascales J et al (1996) Molecular dynamics simulation of a charged biological membrane. J Chem Phys 104:2713–2720
Marrink SJ, De Vries AH, Tieleman DP (2009) Lipids on the move: simulations of membrane pores, domains, stalks and curves. Biochim Biophys Acta (BBA) Biomembr 1788:149–168
Metzler R, Jeon J-H, Cherstvy A (2016) Non-Brownian diffusion in lipid membranes: experiments and simulations. Biochim Biophys Acta (BBA) Biomembr 1858:2451–2467
Miettinen MS et al (2009) Ion dynamics in cationic lipid bilayer systems in saline solutions. J Phys Chem B 113:9226–9234
Mori T et al (2016) Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochim Biophys Acta (BBA) Biomembr 1858:1635–1651
Mukhopadhyay P, Monticelli L, Tieleman DP (2004) Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys J 86:1601–1609
Nademi Y et al (2014) Molecular dynamics simulations and free energy profile of paracetamol in DPPC and DMPC lipid bilayers. J Chem Sci 126:637–647
Ortega-Guerrero A, Espinosa-Duran JM, Velasco-Medina J (2016) TRPV1 channel as a target for cancer therapy using CNT-based drug delivery systems. Eur Biophys J 45:423–433
Paloncýová MT, Berka K, Otyepka M (2013) Molecular insight into affinities of drugs and their metabolites to lipid bilayers. J Phys Chem B 117:2403–2410
Pandit SA, Berkowitz ML (2002) Molecular dynamics simulation of dipalmitoylphosphatidylserine bilayer with Na+ counterions. Biophys J 82:1818–1827
Pant P, Afshan Shaikh S, Jayaram B (2017) Design and characterization of symmetric nucleic acids via molecular dynamics simulations. Biopolymers. https://doi.org/10.1002/bip.23002
Patra MC et al (2014) Molecular dynamics simulation of human serum paraoxonase 1 in DPPC bilayer reveals a critical role of transmembrane helix H1 for HDL association. Eur Biophys J 43:35–51
Petrache HI et al (2004) Structure and fluctuations of charged phosphatidylserine bilayers in the absence of salt. Biophys J 86:1574–1586
Pliquett U et al (2007) High electrical field effects on cell membranes. Bioelectrochemistry 70:275–282
Pohar A, Likozar B (2014) Dissolution, nucleation, crystal growth, crystal aggregation, and particle breakage of amlodipine salts: modeling crystallization kinetics and thermodynamic equilibrium, scale-up, and optimization. Ind Eng Chem Res 53:10762–10774
Polyansky AA et al (2005) Role of lipid charge in organization of water/lipid bilayer interface: insights via computer simulations. J Phys Chem B 109:15052–15059
Ramirez P et al (2006) Theoretical description of the ion transport across nanopores with titratable fixed charges. Cell Biochem Biophys 44:287–312
Riedl S, Zweytick D, Lohner K (2011) Membrane-active host defense peptides–challenges and perspectives for the development of novel anticancer drugs. Chem Phys Lipid 164:766–781
Riedl S et al (2015) Human lactoferricin derived di-peptides deploying loop structures induce apoptosis specifically in cancer cells through targeting membranous phosphatidylserine. Biochim Biophys Acta (BBA) Biomembr 1848:2918–2931
Romo TD et al (2011) Membrane binding of an acyl-lactoferricin B antimicrobial peptide from solid-state NMR experiments and molecular dynamics simulations. Biochim Biophys Acta (BBA) Biomembr 1808:2019–2030
Saiz L, Bandyopadhyay S, Klein ML (2002) Towards an understanding of complex biological membranes from atomistic molecular dynamics simulations. Biosci Rep 22:151–173
Scrima M et al (2014) Structural features of the C8 antiviral peptide in a membrane-mimicking environment. Biochim Biophys Acta (BBA) Biomembr 1838:1010–1018
Shimokawa N et al (2010) Phase separation of a mixture of charged and neutral lipids on a giant vesicle induced by small cations. Chem Phys Lett 496:59–63
Siontorou CG et al (2017) Artificial lipid membranes: past, present, and future. Membranes 7:38
Slochower DR et al (2014) Counterion-mediated pattern formation in membranes containing anionic lipids. Adv Coll Interface Sci 208:177–188
Sun Y et al (2008) The bound states of amphipathic drugs in lipid bilayers: study of curcumin. Biophys J 95:2318–2324
Sun L et al (2018) Coarse-grained molecular dynamics simulation of interactions between cyclic lipopeptide Bacillomycin D and cell membranes. Mol Simul 44:364–376
Van Der Spoel D et al (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Verma A, Stellacci F (2010) Effect of surface properties on nanoparticle–cell interactions. Small 6:12–21
Wagner AJ, May S (2007) Electrostatic interactions across a charged lipid bilayer. Eur Biophys J 36:293–303
Wang H, Meng F (2016) Concentration effect of cimetidine with POPC bilayer: a molecular dynamics simulation study. Mol Simul 42:1292–1297
Wu J, Morikis D (2006) Molecular thermodynamics for charged biomacromolecules. Fluid Phase Equilib 241:317–333
Yang L, Tucker IG, Østergaard J (2011) Effects of bile salts on propranolol distribution into liposomes studied by capillary electrophoresis. J Pharm Biomed Anal 56:553–559
Yousefpour A et al (2013) Molecular dynamics simulation of nonsteroidal antiinflammatory drugs, naproxen and relafen, in a lipid bilayer membrane. Int J Quantum Chem 113:1919–1930
Yousefpour A et al (2015) Interaction of PEGylated anti-hypertensive drugs, amlodipine, atenolol and lisinopril with lipid bilayer membrane: a molecular dynamics simulation study. Biochim Biophys Acta (BBA) Biomembr 1848:1687–1698
Yousefpour A et al (2017) Combination of anti-hypertensive drugs: a molecular dynamics simulation study. J Mol Model 23:158
Zhang Y et al (2010) Targeting therapy with mitosomal daunorubicin plus amlodipine has the potential to circumvent intrinsic resistant breast cancer. Mol Pharm 8:162–175
Zhang T et al (2018) Characterizing the interactions of two lipid modifications with lipid rafts: farnesyl anchors vs. palmitoyl anchors. Eur Biophys J 47:19–30
Zhou F, Schulten K (1995) Molecular dynamics study of a membrane–water interface. J Phys Chem 99:2194–2207
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
FigSM. 1
Drug-drug intermolecular distance for the simulation systems. (TIFF 1693 kb)
Rights and permissions
About this article
Cite this article
Yousefpour, A., Amjad-Iranagh, S., Goharpey, F. et al. Effect of drug amlodipine on the charged lipid bilayer cell membranes DMPS and DMPS + DMPC: a molecular dynamics simulation study. Eur Biophys J 47, 939–950 (2018). https://doi.org/10.1007/s00249-018-1317-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-018-1317-z