Abstract
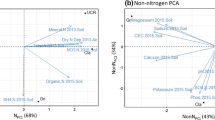
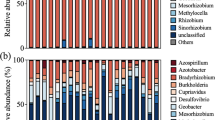
Long-term soil age gradients are useful model systems to study how changes in nutrient limitation shape communities of plant root mutualists because they represent strong natural gradients of nutrient availability, particularly of nitrogen (N) and phosphorus (P). Here, we investigated changes in the dinitrogen (N2)-fixing bacterial community composition and diversity in nodules of a single host legume (Acacia rostellifera) across the Jurien Bay chronosequence, a retrogressive 2 million-year-old sequence of coastal dunes representing an exceptionally strong natural soil fertility gradient. We collected nodules from plants grown in soils from five chronosequence stages ranging from very young (10s of years; associated with strong N limitation for plant growth) to very old (> 2,000,000 years; associated with strong P limitation), and sequenced the nifH gene in root nodules to determine the composition and diversity of N2-fixing bacterial symbionts. A total of 335 unique nifH gene operational taxonomic units (OTUs) were identified. Community composition of N2-fixing bacteria within nodules, but not diversity, changed with increasing soil age. These changes were attributed to pedogenesis-driven shifts in edaphic conditions, specifically pH, exchangeable manganese, resin-extractable phosphate, nitrate and nitrification rate. A large number of common N2-fixing bacteria genera (e.g. Bradyrhizobium, Ensifer, Mesorhizobium and Rhizobium) belonging to the Rhizobiaceae family (α-proteobacteria) comprised 70% of all raw sequences and were present in all nodules. However, the oldest soils, which show some of the lowest soil P availability ever recorded, harboured the largest proportion of unclassified OTUs, suggesting a unique set of N2-fixing bacteria adapted to extreme P limitation. Our results show that N2-fixing bacterial composition varies strongly during long-term ecosystem development, even within the same host, and therefore rhizobia show strong edaphic preferences.



Similar content being viewed by others
References
Kiers ET, Denison RF (2008) Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu Rev Ecol Evol Syst 39:215–236. https://doi.org/10.1146/annurev.ecolsys.39.110707.173423
Sprent J, Ardley J, James E (2013) From North to South: a latitudinal look at legume nodulation processes. S Afr J Bot 89:31–41. https://doi.org/10.1016/j.sajb.2013.06.011
The current taxonomy of rhizobia. NZ Rhizobia website. http://www.rhizobia.co.nz/taxonomy/rhizobia Last updated: X Jan, 2016. [Accessed on 20th of Jan, 2016]. (2016). Accessed 12/06/2015
Thrall PH, Slattery JF, Broadhurst LM, Bickford S (2007) Geographic patterns of symbiont abundance and adaptation in native Australian Acacia–rhizobia interactions. J Ecol 95(5):1110–1122. https://doi.org/10.1111/j.1365-2745.2007.01278.x
Thrall PH, Burdon JJ, Woods MJ (2000) Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian legumes: interactions within and between genera. J Appl Ecol 37(1):52–65. https://doi.org/10.1046/j.1365-2664.2000.00470.x
Sherry SP (1971) The Black Wattle (Acacia mearnsii De Wild.). University of Natal Press, Pietermaritzburg
Boddey R, De Oliveira O, Urquiaga S, Reis V, De Olivares F, Baldani V, Döbereiner J (1995) Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil 174. https://doi.org/10.1007/978-94-011-0053-3_9
Heath KD, Tiffin P (2007) Context dependence in the coevolution of plant and rhizobial mutualists. Proc R Soc B: Biol Sc 274(1620):1905–1912. https://doi.org/10.1098/rspb.2007.0495
Weese DJ, Heath KD, Dentinger BTM, Lau JA (2015) Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69(3):631–642. https://doi.org/10.1111/evo.12594
Simonsen AK, Han S, Rekret P, Rentschler CS, Heath KD, Stinchcombe JR (2015) Short-term fertilizer application alters phenotypic traits of symbiotic nitrogen fixing bacteria. PeerJ 3:e1291. https://doi.org/10.7717/peerj.1291
VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam SJ, Mohn WW (2015) Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9(11):2435–2441. https://doi.org/10.1038/ismej.2015.54
Hollowell A, Regus J, Gano K, Bantay R, Centeno D, Pham J, Lyu J, Moore D, Bernardo A, Lopez G (2016) Epidemic spread of symbiotic and non-symbiotic Bradyrhizobium genotypes across California. Microb Ecol 71(3):700–710. https://doi.org/10.1007/s00248-015-0685-5
Neuhauser C, Fargione JE (2004) A mutualism-parasitism continuum model and its application to plant-mycorrhizae interactions. Ecol Model 177(3–4):337–352. https://doi.org/10.1016/j.ecolmodel.2004.02.010
Van Cauwenberghe J, Michiels J, Honnay O (2015) Effects of local environmental variables and geographical location on the genetic diversity and composition of Rhizobium leguminosarum nodulating Vicia cracca populations. Soil Biol Biochem 90:71–79. https://doi.org/10.1016/j.soilbio.2015.08.001
Denison RF, Kiers ET (2004) Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Ecol 237(2):187–193. https://doi.org/10.1111/j.1574-6968.2004.tb09695.x
Barrett LG, Bever JD, Bissett A, Thrall PH (2015) Partner diversity and identity impacts on plant productivity in Acacia–rhizobial interactions. J Ecol 103(1):130–142. https://doi.org/10.1111/1365-2745.12336
Ferreira TC, Aguilar JV, Souza LA, Justino GC, Aguiar LF, Camargos LS (2016) pH effects on nodulation and biological nitrogen fixation in Calopogonium mucunoides. Braz J Bot 39(4):1015–1020. https://doi.org/10.1007/s40415-016-0300-0
Slattery JF, Coventry DR (1993) Variation of soil populations of Rhizobium leguminosarum bv. Trifolii and the occurrence of inoculant rhizobia in nodules of subterranean clover after pasture renovation in north-eastern Victoria. Soil Biol Biochem 25(12):1725–1730. https://doi.org/10.1016/0038-0717(93)90176-C
Vuong HB, Thrall PH, Barrett LG (2016) Host species and environmental variation can influence rhizobial community composition. J Ecol 105:540–548. https://doi.org/10.1111/1365-2745.12687
Walker LR, Wardle DA, Bardgett RD, Clarkson BD (2010) The use of chronosequences in studies of ecological succession and soil development. J Ecol 98(4):725–736. https://doi.org/10.1111/j.1365-2745.2010.01664.x
Krüger M, Teste FP, Laliberté E, Lambers H, Coghlan M, Zemunik G, Bunce M (2015) The rise and fall of arbuscular mycorrhizal fungal diversity during ecosystem retrogression. Mol Ecol 24(19):4912–4930. https://doi.org/10.1111/mec.13363
Dickie IA, Martínez-García LB, Koele N, Grelet G-A, Tylianakis JM, Peltzer DA, Richardson SJ (2013) Mycorrhizas and mycorrhizal fungal communities throughout ecosystem development. Plant Soil 367(1–2):11–39. https://doi.org/10.1007/s11104-013-1609-0
Albornoz FE, Lambers H, Turner BL, Teste FP, Laliberté E (2016) Shifts in symbiotic associations in plants capable of forming 2 multiple root symbioses across a long-term soil chronosequence. Ecol Evol 6(8):2368–2377. https://doi.org/10.1002/ece3.2000
Turner BL, Laliberté E (2015) Soil development and nutrient availability along a 2 million-year coastal dune chronosequence under species-rich Mediterranean shrubland in Southwestern Australia. Ecosystems 18(2):287–309. https://doi.org/10.1007/s10021-014-9830-0
Vitousek PM (2004) Nutrient cycling and limitation: Hawai’i as a model system. Princeton University Press, USA
Crews TE, Kitayama K, Fownes JH, Riley RH, Herbert DA, Mueller-Dombois D, Vitousek PM (1995) Changes in soil phosphorus fractions and ecosystem dynamics across a long chronosequence in Hawaii. Ecol 76(5):1407–1424. https://doi.org/10.2307/1938144
Peltzer DA, Wardle DA, Allison VJ, Baisden WT, Bardgett RD, Chadwick OA, Condron LM, Parfitt RL, Porder S, Richardson SJ, Turner BL, Vitousek PM, Walker J, Walker LR (2010) Understanding ecosystem retrogression. Ecol Monogr 80(4):509–529. https://doi.org/10.1890/09-1552.1
Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD (2015) Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol 205(4):1525–1536. https://doi.org/10.1111/nph.13208
Martínez-García LB, Richardson SJ, Tylianakis JM, Peltzer DA, Dickie IA (2015) Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytol 205(4):1565–1576. https://doi.org/10.1111/nph.13226
Laliberté E, Turner BL, Costes T, Pearse SJ, Wyrwoll KH, Zemunik G, Lambers H (2012) Experimental assessment of nutrient limitation along a 2-million-year dune chronosequence in the south-western Australia biodiversity hotspot. J Ecol 100(3):631–642. https://doi.org/10.1111/j.1365-2745.2012.01962.x
McArthur, WM & Bettenay, E (1974) Development and Distribution of Soils of the Swan Coastal Plain, Western Australia. CSIRO, Australia.
Odum EP (1969) The strategy of ecosystem development. In: Ndubisi FO (ed) The ecological design and planning reader. Island Press/Center for Resource Economics, Washington, DC, pp 203–216. doi:https://doi.org/10.5822/978-1-61091-491-8_20
Vitousek PM, Farrington H (1997) Nutrient limitation and soil development: experimental test of a biogeochemical theory. Biogeochem 37(1):63–75. https://doi.org/10.1023/A:1005757218475
Kitayama K, Mueller-Dombois D (1995) Vegetation changes along gradients of long-term soil development in the Hawaiian montane rainforest zone. Plant Ecol 120(1):1–20. https://doi.org/10.1007/BF00033454
Richardson SJ, Peltzer DA, Allen RB, McGlone MS, Parfitt RL (2004) Rapid development of phosphorus limitation in temperate rainforest along the Franz Josef soil chronosequence. Oecologia 139(2):267–276. https://doi.org/10.1007/s00442-004-1501-y
Laliberté E, Zemunik G, Turner BL (2014) Environmental filtering explains variation in plant diversity along resource gradients. Science 345(6204):1602–1605. https://doi.org/10.1126/science.1256330
Zemunik G, Turner BL, Lambers H, Laliberté E (2016) Increasing plant species diversity and extreme species turnover accompany declining soil fertility along a long-term chronosequence in a biodiversity hotspot. J Ecol 104(3):792–805. https://doi.org/10.1111/1365-2745.12546
Albornoz FE, Teste FP, Lambers H, Bunce M, Murray DC, White NE, Laliberté E (2016) Changes in ectomycorrhizal fungal community composition and declining diversity along a 2-million year soil chronosequence. Mol Ecol 25(19):4919–4929. https://doi.org/10.1111/mec.13778
Raven JA (2012) Protein turnover and plant RNA and phosphorus requirements in relation to nitrogen fixation. Plant Sc 188:25–35. https://doi.org/10.1016/j.plantsci.2012.02.010
Sprent JI, Raven JA (1985) Evolution of nitrogen-fixing symbioses. Proc R Soc Edinb B Biol Sci 85(3–4):215–237. https://doi.org/10.1017/S0269727000004036
Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5(7):539–554. https://doi.org/10.1046/j.1462-2920.2003.00451.x
Gaby JC, Buckley DH (2011) A global census of nitrogenase diversity. Environ Microbiol 13(7):1790–1799. https://doi.org/10.1111/j.1462-2920.2011.02488.x
Groves RH (ed) (1994) Australian vegetation2nd edn. Melbourne, Cambridge University Press
Thrall PH, Bever JD, Slattery JF (2008) Rhizobial mediation of Acacia adaptation to soil salinity: evidence of underlying trade-offs and tests of expected patterns. J Ecol 96(4):746–755. https://doi.org/10.1111/j.1365-2745.2008.01381.x
Graham PH (1992) Stress tolerance in Rhizobium and Bradyrhizobium, and nodulation under adverse soil conditions. Can J Microbiol 38(6):475–484. https://doi.org/10.1139/m92-079
Lowendorf HS, Alexander M (1983) Identification of Rhizobium phaseoli strains that are tolerant or sensitive to soil acidity. Appl Environ Microbiol 45(3):737–742
McArthur WM, Bettenay E (1960) The development and distribution of the soils of the Swan Coastal Plain, Western Australia. Soil Publication CSIRO, Australia 16
Zemunik G, Turner BL, Lambers H, Laliberté E (2015) Diversity of plant nutrient-acquisition strategies increases during long-term ecosystem development. Nature Plants 1(5):15050. https://doi.org/10.1038/nplants.2015.50
Enright NJ, Lamont BB, Miller BP (2005) Anomalies in grasstree fire history reconstructions for south-western Australian vegetation. Austral Ecol 30(6):668–673. https://doi.org/10.1111/j.1442-9993.2005.01509.x
Enright NJ, Fontaine JB, Lamont BB, Miller BP, Westcott VC (2014) Resistance and resilience to changing climate and fire regime depend on plant functional traits. J Ecol 102(6):1572–1581
Hoque MS, Broadhurst LM, Thrall PH (2011) Genetic characterisation of root nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across southeastern Australia. Int J Syst Evol Microbiol 61:299–309. https://doi.org/10.1099/ijs.0.021014-0
Birnbaum C, Bissett A, Thrall PH, Leishman MR (2016) Nitrogen-fixing bacterial communities in invasive legume nodules and associated soils are similar across introduced and native range populations in Australia. J Biogeogr 43(8):1631–1644. https://doi.org/10.1111/jbi.12752
Hill Y (2015) Investigation of the symbiotic associations of Acacia ligulata Benth. and Acacia tetragonophylla F.Muell: the potential for use in the rehabilitation of excavated sites at Shark Bay Salt Pty. Ltd. PhD Thesis. Murdoch University, Perth, PerthAustralia
Hayes P, Turner BL, Lambers H, Laliberté E (2014) Foliar nutrient concentrations and resorption efficiency in plants of contrasting nutrient-acquisition strategies along a 2-million-year dune chronosequence. J Ecol 102(2):396–410. https://doi.org/10.1111/1365-2745.12196
Zehr JP, McReynolds LA (1989) Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55(10):2522–2526
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998. https://doi.org/10.1038/nmeth.2604
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217
Gaby JC, Buckley DH (2014) A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014:bau001
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32(4):1363–1371. https://doi.org/10.1093/nar/gkh293
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26(1):32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x
Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Springer, New York
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Peter Solymos P, Stevens MHH, Szoecs E, Wagner H (2017) Vegan: community ecology package. R package version 2:4–3 http://CRAN.R-project.org/package=vegan
Cáceres MD, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90(12):3566–3574. https://doi.org/10.1890/08-1823.1
Venables WN, Ripley BD (2002) Modern applied statistics with SFourth edn. Springer, New York
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Hope RM (2013) Rmisc: Ryan miscellaneous. R package version 1:5
Navarro DJ (2015) Learning statistics with R: a tutorial for psychology students and other beginners. (Version 0.5). University of Adelaide, Adelaide, Australia
Taylor SR, McLennan SM (1985) The continental crust: its composition and evolution, vol 1. Blackwell Press, Oxford
Chen CR, Hou EQ, Condron LM, Bacon G, Esfandbod M, Olley J, Turner BL (2015) Soil phosphorus fractionation and nutrient dynamics along the Cooloola coastal dune chronosequence, southern Queensland, Australia. Geoderma 257-258(supplement C):4–13. https://doi.org/10.1016/j.geoderma.2015.04.027
Png GK, Turner BL, Albornoz FE, Hayes PE, Lambers H, Laliberté E (2017) Greater root phosphatase activity in nitrogen-fixing rhizobial but not actinorhizal plants with declining phosphorus availability. J Ecol n/a:n/a 105:1246–1255. https://doi.org/10.1111/1365-2745.12758
Krasova-Wade T, Diouf O, Ndoye I, Sall CE, Braconnier S, Neyra M (2006) Water-condition effects on rhizobia competition for cowpea nodule occupancy. Afr J Biotechnol 5(16):1457–1463
Brockwell J, Pilka A, Holliday R (1991) Soil pH is a major determinant of the numbers of naturally occurring Rhizobium meliloti in non-cultivated soils in central New South Wales. Austr J Exp Agr 31(2):211–219. https://doi.org/10.1071/EA9910211
West S, Kiers ET, Pen I, Denison R (2002) Sanctions and mutualism stability: when should less beneficial mutualists be tolerated? J Evol Biol 15(5):830–837. https://doi.org/10.1046/j.1420-9101.2002.00441.x
Futuyma DJ, Moreno G (1988) The evolution of ecological specialization. Ann Rev Ecol System 19:207–233. https://doi.org/10.1146/annurev.es.19.110188.001231
Cooper VS, Lenski RE (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407(6805):736–739. https://doi.org/10.1038/35037572
Vinuesa P, Neumann-Silkow F, Pacios-Bras C, Spaink HP, Martínez-Romero E, Werner D (2003) Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol plant-microbe inter 16(2):159–168. https://doi.org/10.1094/MPMI.2003.16.2.159
Martyniuk S, Oron J, Martyniuk M (2005) Diversity and numbers of root-nodule bacteria [rhizobia] in Polish soils. Acta Soc Bot Pol 74(1):83–86
Wielbo J, Kidaj D, Koper P, Kubik-Komar A, Skorupska A (2012) The effect of biotic and physical factors on the competitive ability of Rhizobium leguminosarum. Open Life Sci 7(1):13–24
Provorov NA, Vorobyov NI (2006) Interplay of Darwinian and frequency-dependent selection in the host-associated microbial populations. Theor Pop Biol 70(3):262–272. https://doi.org/10.1016/j.tpb.2006.06.002
Sachs J, Kembel S, Lau A, Simms E (2009) In situ phylogenetic structure and diversity of wild Bradyrhizobium communities. Appl Environ Microbiol 75(14):4727–4735
Turk D, Keyser HH (1992) Rhizobia that nodulate tree legumes: specificity of the host for nodulation and effectiveness. Can J Microbiol 38(6):451–460. https://doi.org/10.1139/m92-076
Lodeiro AR, Favelukes G (1999) Early interactions of Bradyrhizobium japonicum and soybean roots: specificity in the process of adsorption. Soil Biol Biochem 31(10):1405–1411. https://doi.org/10.1016/S0038-0717(99)00058-9
Lieven-Antoniou C, Whittam T (1997) Specificity in the symbiotic association of Lotus corniculatus and Rhizobium loti from natural populations. Mol Ecol 6(7):629–639. https://doi.org/10.1046/j.1365-294X.1997.00224.x
Walker T, Syers J (1976) The fate of phosphorus during pedogenesis. Geoderma 15(1):1–19. https://doi.org/10.1016/0016-7061(76)90066-5
Batterman SA, Wurzburger N, Hedin LO (2013) Nitrogen and phosphorus interact to control tropical symbiotic N2 fixation: a test in Inga punctata. J Ecol 101(6):1400–1408. https://doi.org/10.1111/1365-2745.12138
Acknowledgements
We thank Yvette Hill who provided comments and information of rhizobia nodulating A. rostellifera. We appreciate the cooperation by the Western Australian Department of Parks and Wildlife (DPaW) with acquiring permits to sample the field soils. We are grateful for the hard work of Yuphin Khentry and Ghulam Abbas during the soil collections and glasshouse measurements.
Funding
This research was made possible by research grants from the Australian Research Council (DE120100352 and DP130100016) and the Hermon Slade Foundation to E.L.
Author information
Authors and Affiliations
Contributions
FPT and EL designed the experiment. FPT supervised and maintained the experiment. CB conducted the DNA extractions. CB performed statistical analyses with assistance from all authors. AB conducted the bioinformatics work. CB led the writing of the manuscript and all authors contributed to revisions.
Corresponding author
Electronic Supplementary Material
Online resource 1
List of 30 soil variables, sampling levels and methods used in dbRDA analyses. (XLSX 13 kb)
Online resource 2
Means and ± S.E. of 30 soil variables used in the dbRDA analysis. (XLSX 13 kb)
ESM 1
(DOCX 21 kb)
Online resource 5
Phylogenetic tree showing the 335 OTUs based on nifH gene amplified from A. rostellifera nodules’ DNA after growing in the greenhouse in soils collected across five chronosequence stages in Jurien Bay, Western Australia. The tree is based on the neighbour joining tree of Gaby et al. (2014) to which 335 OTU sequences were added using the ARB parsimony tool. Three phylogenetically distinct clusters are shown. In blue are marked the 12 most common OTUs based on Table 2. In green are marked OTUs that associated significantly with dune stage 5 (the oldest) based on Table 3. (attached separately) (PDF 50 kb)
Rights and permissions
About this article
Cite this article
Birnbaum, C., Bissett, A., Teste, F.P. et al. Symbiotic N2-Fixer Community Composition, but Not Diversity, Shifts in Nodules of a Single Host Legume Across a 2-Million-Year Dune Chronosequence. Microb Ecol 76, 1009–1020 (2018). https://doi.org/10.1007/s00248-018-1185-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1185-1