Abstract
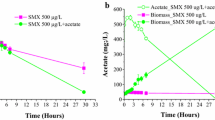
Over-the-counter pharmaceutical compounds can serve as microbial substrates in wastewater treatment processes as well as in the environment. The metabolic pathways and intermediates produced during their degradation, however, are poorly understood. In this study, we investigate an anaerobic wastewater community that metabolizes naproxen via demethylation. Enriched cultures, established from anaerobic digester inocula receiving naproxen as the sole carbon source, transformed naproxen to 6-O-desmethylnaproxen (DMN) within 22 days. Continual enrichment and culture transfer resulted in consistent demethylation of naproxen with no loss of DMN observed. Methane was generated at 0.83 mmol per 1 mmol transformed naproxen. In addition to naproxen, the consortium readily demethylated syringic acid and vanillic acid. DNA analysis revealed a community of acetogenic bacteria and syntrophic acetate oxidizing archaea. Combined with the biotransformation data, this suggests the enriched consortium performs aromatic O-demethylation through a syntrophic relationship between specific acetogens, acetate oxidizers, and methanogens. The proposed model of carbon transfer through the anaerobic food web highlights the significance of linked community interactions in the anaerobic transformation of aromatic O-methyl compounds such as naproxen.





Similar content being viewed by others
References
Brozinski J-M, Lahti M, Meierjohann A et al (2013) The anti-inflammatory drugs diclofenac, naproxen and ibuprofen are found in the bile of wild fish caught downstream of a wastewater treatment plant. Environ Sci Technol 47:342–348. https://doi.org/10.1021/es303013j
Gross B, Montgomery-Brown J, Naumann A, Reinhard M (2004) Occurrence and fate of pharmaceuticals and alkylphenol ethoxylate metabolites in an effluent-dominated river and wetland. Environ Toxicol Chem 23:2074–2083
Tixier C, Singer HP, Oellers S, Müller SR (2003) Occurrence and fate of carbamazepine, clofibric acid, diclofenac, ibuprofen, ketoprofen, and naproxen in surface waters. Environ Sci Technol 37:1061–1068. https://doi.org/10.1021/es025834r
Goldstein M, Shenker M, Chefetz B (2014) Insights into the uptake processes of wastewater-borne pharmaceuticals by vegetables. Environ Sci Technol 48:5593–5600. https://doi.org/10.1021/es5008615
Carballa M, Fink G, Omil F et al (2008) Determination of the solid–water distribution coefficient (K d) for pharmaceuticals, estrogens and musk fragrances in digested sludge. Water Res 42:287–295. https://doi.org/10.1016/j.watres.2007.07.012
Maoz A, Chefetz B (2010) Sorption of the pharmaceuticals carbamazepine and naproxen to dissolved organic matter: role of structural fractions. Water Res 44:981–989. https://doi.org/10.1016/j.watres.2009.10.019
Quintana J, Weiss S, Reemtsma T (2005) Pathways and metabolites of microbial degradation of selected acidic pharmaceutical and their occurrence in municipal wastewater treated by a membrane bioreactor. Water Res 39:2654–2664. https://doi.org/10.1016/j.watres.2005.04.068
Lahti M, Oikari A (2011) Microbial transformation of pharmaceuticals naproxen, bisoprolol, and diclofenac in aerobic and anaerobic environments. Arch Environ Contam Toxicol 61:202–210. https://doi.org/10.1007/s00244-010-9622-2
Daniel SL, Keith ES, Yang H et al (1991) Utilization of methoxylated aromatic compounds by the acetogen Clostridium thermoaceticum: expression and specificity of the co-dependent O-demethylating activity. Biochem Biophys Res Commun 180:416–422. https://doi.org/10.1016/S0006-291X(05)81309-9
Phelps CD, Young LY (1997) Microbial metabolism of the plant phenolic compounds ferulic and syringic acids under three anaerobic conditions. Microb Ecol 33:206–215
Owen W, Stuckey D, Healy Jr J et al (1979) Bioassay for monitoring biochemical methane potential and anaerobic toxicity. Water Res 13:485–492. https://doi.org/10.1016/0043-1354(79)90043-5
Andersen JV, Hansen SH (1992) Simultaneous determination of (R)- and (S)-naproxen and (R)- and (S)-6-O-desmethylnaproxen by high-performance liquid chromatography on a Chiral-AGP column. J Chromatogr 577:362–365
Healy JB, Young LY (1979) Anaerobic biodegradation of eleven aromatic compounds to methane. Appl Environ Microbiol 38:84–89
Zinder SH, Anguish T, Cardwell SC (1984) Selective inhibition by 2-bromoethanesulfonate of methanogenesis from acetate in a thermophilic anaerobic digestor. Appl Environ Microbiol 47:1343–1345
Kaiser J-P, Hanselmann KW (1982) Fermentative metabolism of substituted monoaromatic compounds by a bacterial community from anaerobic sediments. Arch Microbiol 133:185–194. https://doi.org/10.1007/BF00414999
Parks GS, Huffman HM (1932) The free energies of some organic compounds. Chemical Catalog Company, Incorporated
Laviska DA, Guan C, Emge TJ et al (2014) Addition of C–C and C–H bonds by pincer-iridium complexes: a combined experimental and computational study. Dalton Trans 43:16354–16365. https://doi.org/10.1039/C4DT02043J
Leach ST, Lui K, Naing Z et al (2015) Multiple opportunistic pathogens, but not pre-existing inflammation, may be associated with necrotizing enterocolitis. Dig Dis Sci 60:3728–3734. https://doi.org/10.1007/s10620-015-3830-6
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
O’Connor OA, Rivera MD, Young LY (1989) Toxicity and biodegradation of phthalic acid esters under methanogenic conditions. Environ Toxicol Chem 8:569–576
Bache R, Pfennig N (1981) Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol 130:255–261. https://doi.org/10.1007/BF00459530
Frazer AC, Young LY (1985) A gram-negative anaerobic bacterium that utilizes O-methyl substituents of aromatic acids. Appl Environ Microbiol 49:1345–1347
Frazer AC (1994) O-demethylation and other transformations of aromatic compounds by acetogenic bacteria. In: Drake HL (ed) Acetogenesis. Springer US, pp 445–483
Berman MH, Frazer AC (1992) Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl Environ Microbiol 58:925–931
Balk M, Mehboob F, van Gelder AH et al (2010) (Per)chlorate reduction by an acetogenic bacterium, Sporomusa sp., isolated from an underground gas storage. Appl Microbiol Biotechnol 88:595–603. https://doi.org/10.1007/s00253-010-2788-8
Muller V, Frerichs J (2013) Acetogenic bacteria. In eLS. John Wiley & Sons, Ltd: Chichester. https://doi.org/10.1002/9780470015902.a0020086.pub2
Garcia J-L, Ollivier B, Whitman WB (2006) The order methanomicrobiales. In: Dr MDP, Falkow S, Rosenberg E et al (eds) The prokaryotes. Springer, New York, pp 208–230
Oren A (2014) The family methanobacteriaceae. In: Rosenberg E, DeLong EF, Lory S et al (eds) The prokaryotes. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 165–193
Bitton G (2005) Wastewater microbiology. John Wiley & Sons, Hoboken
Zinder SH, Koch M (1984) Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Arch Microbiol 138:263–272. https://doi.org/10.1007/BF00402133
Barker HA (1936) On the biochemistry of the methane fermentation. Arch Für Mikrobiol 7:404–419. https://doi.org/10.1007/BF00407413
Karakashev D, Batstone DJ, Trably E, Angelidaki I (2006) Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol 72:5138–5141. https://doi.org/10.1128/AEM.00489-06
Westerholm M, Roos S, Schnürer A (2010) Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol Lett. https://doi.org/10.1111/j.1574-6968.2010.02023.x
Acknowledgements
The authors gratefully acknowledge Tom Villani and the labs of Dr. James Simon and Dr. Alan Goldman for the analysis support. The nucleotide sequence data reported are available in the NCBI Sequence Read Archive under the BioSample accession numbers SAMN07237492 and SAMN07237493.
Funding
This project was supported by the USDA National Institute of Food and Agriculture Hatch Multistate project 1007899 through the New Jersey Agricultural Experiment Station, Hatch Multistate NJ07212. Sarah Wolfson was supported by a US National Science Foundation Fuels IGERT from Rutgers University (NSF DGE 0903675).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
(DOCX 355kb)
Rights and permissions
About this article
Cite this article
Wolfson, S.J., Porter, A.W., Campbell, J.K. et al. Naproxen Is Transformed Via Acetogenesis and Syntrophic Acetate Oxidation by a Methanogenic Wastewater Consortium. Microb Ecol 76, 362–371 (2018). https://doi.org/10.1007/s00248-017-1136-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1136-2