Abstract
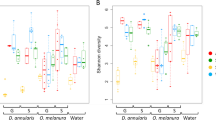
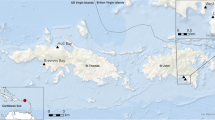
Damselfishes of the genus Stegastes are some of the most abundant fish inhabiting shallow reef habitats. Although Stegastes play an important role in promoting primary production in reef ecosystems, their territories are known to favor the occurrence of bacterial pathogens. However, no studies so far have examined the microbiome composition of Caribbean Stegastes species. Here, we characterized the skin bacterial microbiome of juveniles and adults of Stegastes leucostictus and Stegastes adustus collected from La Parguera, Puerto Rico, and St. Thomas, US Virgin Islands. Our results showed that bacterial communities residing on the skin of Stegastes were related to taxonomy, environment and ontogeny. These results are in line with previous knowledge on the dynamics of fish skin microbiomes.



Similar content being viewed by others
References
Allen GR (1991) Damselfishes of the world. Mergus, Melle
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Buchmann K (1999) Immune mechanisms in fish skin against monogeneans—a model. Folia Parasitol 46:1–9
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581. https://doi.org/10.1038/nmeth.3869(https://www.nature.com/articles/nmeth.3869#supplementary-information)
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Nat Acad Sci 108:4516. https://doi.org/10.1073/pnas.1000080107
Carlson JM, Leonard AB, Hyde ER, Petrosino JF, Primm TP (2017) Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect Drug Resist 10:143–154. https://doi.org/10.2147/idr.s129055
Casey JM, Ainsworth TD, Choat JH, Connolly SR (2014) Farming behaviour of reef fishes increases the prevalence of coral disease associated microbes and black band disease. Proc Biol Sci 281:20141032–20141032. https://doi.org/10.1098/rspb.2014.1032
Chiarello M, Villéger S, Bouvier C, Bettarel Y, Bouvier T (2015) High diversity of skin-associated bacterial communities of marine fishes is promoted by their high variability among body parts, individuals and species. FEMS Microb Ecol 91:fiv06
Chiarello M, Auguet J-C, Bettarel Y, Bouvier C, Claverie T, Graham NAJ, Rieuvilleneuve F, Sucré E, Bouvier T, Villéger S (2018) Skin microbiome of coral reef fish is highly variable and driven by host phylogeny and diet. Microbiome 6:147. https://doi.org/10.1186/s40168-018-0530-4
Collyer ML, Sekora DJ, Adams DC (2015) A method for analysis of phenotypic change for phenotypes described by high-dimensional data. Heredity 115:357–365. https://doi.org/10.1038/hdy.2014.75
Doropoulos C, Hyndes GA, Abecasis D, Vergés A (2013) Herbivores strongly influence algal recruitment in both coral- and algal-dominated coral reef habitats. Mar Ecol Prog Ser 486:153–164
Dromard CR, Bouchon-Navaro Y, Cordonnier S, Fontaine M-F, Verlaque M, Harmelin-Vivien M, Bouchon C (2013) Resource use of two damselfishes, Stegastes planifrons and Stegastes adustus, on Guadeloupean reefs (Lesser Antilles): inference from stomach content and stable isotope analysis. J Exp Mar Biol Ecol 440:116–125. https://doi.org/10.1016/j.jembe.2012.12.011
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Edgar RC (2016) UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. https://doi.org/10.1101/081257. bioRxiv: 081257
Emery AR (1973) Comparative ecology and functional osteology of fourteen species of damselfish (Pisces: Pomacentridae) at alligator reef, Florida keys. Bul Mar Sci 23:649–770
Galbraith H, Iwanowicz D, Spooner D, Iwanowicz L, Keller D, Zelanko P, Adams C (2018) Exposure to synthetic hydraulic fracturing waste influences the mucosal bacterial community structure of the brook trout (Salvelinus fontinalis) epidermis. AIMS Microbiol 4:413–427
Gilbert JA, Jansson JK, Knight R (2014) The Earth microbiome project: successes and aspirations. BMC Biol 12:69. https://doi.org/10.1186/s12915-014-0069-1
Gutiérrez L (1998) Habitat selection by recruits establishes local patterns of adult distribution in two species of damselfishes: Stegastes dorsopunicans and S. planifrons. Oecologia 115:268–277. https://doi.org/10.1007/s004420050516
Hess S, Wenger AS, Ainsworth TD, Rummer JL (2015) Exposure of clownfish larvae to suspended sediment levels found on the Great Barrier Reef: impacts on gill structure and microbiome. Sci Rep 5:10561–10561. https://doi.org/10.1038/srep10561
Hixon MA, Brostoff WN (1983) Damselfish as keystone species in reverse: intermediate disturbance and diversity of reef algae. Science 220:511–513. https://doi.org/10.1126/science.220.4596.511
Itzkowitz M (1977) Spatial organization of the Jamaican damselfish community. J Exp Mar Biol Ecol 28:217–241. https://doi.org/10.1016/0022-0981(77)90093-4
Itzkowitz M (1985) Aspects of the population dynamics and reproductive success in the permanently territorial beaugregory damselfish. Mar Behav Physiol 12:57–69. https://doi.org/10.1080/10236248509378633
Jones KMM (2005) The effect of territorial damselfish (family Pomacentridae) on the space use and behaviour of the coral reef fish, Halichoeres bivittatus (Bloch, 1791) (family Labridae). J Exp Mar Biol Ecol 324:99–111. https://doi.org/10.1016/j.jembe.2005.04.009
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl Environ Microbiol 79:5112. https://doi.org/10.1128/aem.01043-13
Larsen AM, Bullard SA, Womble M, Arias CR (2015) Community structure of skin microbiome of gulf killifish, Fundulus grandis, is driven by seasonality and not exposure to oiled sediments in a Louisiana Salt Marsh. Microb Ecol 70:534–544. https://doi.org/10.1007/s00248-015-0578-7
Lassuy DR (1980) Effects of farming behavior by Eupomacentrus Lividus and Hemiglyphidodon Plagiometopon on algal community structure. Bul Mar Sci 30:304–312
Lassuy DR (1984) Diet, intestinal morphology, and nitrogen assimilation efficiency in the damselfish, Stegastes lividus, in Guam. Environ Biol Fish 10:183–193. https://doi.org/10.1007/bf00001125
Legrand TPRA, Catalano SR, Wos-Oxley ML, Stephens F, Landos M, Bansemer MS, Stone DAJ, Qin JG, Oxley APA (2018) The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front Microbiol 8:1. https://doi.org/10.3389/fmicb.2017.02664
Llewellyn MS, McGinnity P, Dionne M, Letourneau J, Thonier F, Carvalho GR, Creer S, Derome N (2016) The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. ISME 10:1280–1284. https://doi.org/10.1038/ismej.2015.189
Lokesh J, Kiron V (2016) Transition from freshwater to seawater reshapes the skin associated microbiota of Atlantic salmon. Sci Rep 6:19707
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Lowrey L, Woodhams DC, Tacchi L, Salinas I (2015) Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol 81:6915. https://doi.org/10.1128/aem.01826-15
Marchesi JR, Ravel J (2015) The vocabulary of microbiome research: a proposal. Microbiome 3:31. https://doi.org/10.1186/s40168-015-0094-5
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Miyake S, Ngugi DK, Stingl U (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol Ecol 24:656–672. https://doi.org/10.1111/mec.13050
Myrberg AA (1972) Ethology of the bicolor damselfish, Eupomacentrus partitus (Pisces: Pomacentridae): a comparative analysis of laboratory and field behaviour. Anim Behav Monogr 5: 197-IN192. https://doi.org/10.1016/0003-3472(72)90002-4
Myrberg AAJ, Thresher RE (1974) Interspecific aggression and its relevance to the concept of territoriality in reef fishes. Am Zool 14:81–96. https://doi.org/10.1093/icb/14.1.81
Nicholson MD, Sikkel PC (2018) Localized defecation in territorial herbivorous fishes. Copeia 106:532–538
Nielsen S, Wilkes Walburn J, Vergés A, Thomas T, Egan S (2017) Microbiome patterns across the gastrointestinal tract of the rabbitfish Siganus fuscescens. PeerJ 5:e3317–e3317. https://doi.org/10.7717/peerj.3317
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Package ‘Vegan’. Community ecology package, Version 2. https://CRAN.R-project.org/package=vegan
Parris DJ, Brooker RM, Morgan MA, Dixson DL, Stewart FJ (2016) Whole gut microbiome composition of damselfish and cardinalfish before and after reef settlement. PeerJ 4:e2412–e2412. https://doi.org/10.7717/peerj.2412
Pratte ZA, Besson M, Hollman RD, Stewart FJ (2018) The gills of reef fish support a distinct microbiome influenced by host-specific factors. Appl Environ Microbiol 84:e00063–e118. https://doi.org/10.1128/aem.00063-18
Reverter M, Sasal P, Tapissier-Bontemps N, Lecchini D, Suzuki M (2017) Characterisation of the gill mucosal bacterial communities of four butterflyfish species: a reservoir of bacterial diversity in coral reef ecosystems. FEMS Microbiol Ecol 93:1. https://doi.org/10.1093/femsec/fix051
Rosado D, Pérez-Losada M, Severino R, Cable J, Xavier R (2019a) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture 500:57–64. https://doi.org/10.1016/j.aquaculture.2018.09.063
Rosado D, Xavier R, Severino R, Tavares F, Cable J, Pérez-Losada M (2019b) Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sc Rep 9:18946. https://doi.org/10.1038/s41598-019-55314-4
Sikkel PC (1998) Competitor intrusions and mate-search tactics in a territorial marine fish. Behav Ecol 9:439–444
Sikkel PC, Fuller CA, Hunte W (2000) Habitat/sex differences in time at cleaning stations and ectoparasite loads in a Caribbean reef fish. Mar Ecol Prog Ser 193:191–199
Sikkel PC, Herzlieb SE, Kramer DL (2005) Compensatory cleaner-seeking behavior following spawning in female yellowtail damselfish. Mar Ecol Prog Ser 296:1–11
Sikkel PC, Kramer DL (2006) Territory revisits reduce intrusion during spawning trips by female yellowtail damselfish, Microspathodon chrysurus. An Behav 71:71–78
Sikkel PC, Nemeth D, McCammon A, Williams EH Jr (2009) Habitat and species differences in prevalence and intensity of Neobenedenia melleni (Monogenea: Capsalidae) on sympatric Caribbean surgeonfishes (Acanthuridae). J Parasitol 95:63–68
Soares MC, Cable J, Lima-Maximino MG, Maximino C, Xavier R (2019) Using fish models to investigate the links between microbiome and social behaviour: the next step for translational microbiome research? Fish Fish 20:640652. https://doi.org/10.1111/faf.12366
Sweatman H (1988) Field evidence that settling coral reef fish larvae detect resident fishes using dissolved chemical cues. J Exp Mar Biol Ecol 124:163–174. https://doi.org/10.1016/0022-0981(88)90170-0
Sweatman HPA (1983) Influence of conspecifics on choice of settlement sites by larvae of two pomacentrid fishes (Dascyllus aruanus and D. reticulatus) on coral reefs. Mar Biol 75:225–229. https://doi.org/10.1007/bf00406006
Sylvain F-É, Derome N (2017) Vertically and horizontally transmitted microbial symbionts shape the gut microbiota ontogenesis of a skin-mucus feeding discus fish progeny. Sci Rep 7:5263. https://doi.org/10.1038/s41598-017-05662-w
Tarnecki AM, Brennan NP, Schloesser RW, Rhoder NR (2019) Shifts in the skin-associated microbiota of hatchery-reared common snook Centropomus undecimalis during acclimation to the wild. Microb Ecol 77:770–781. https://doi.org/10.1007/s00248-018-1252-7
Waldner RE, Robertson DR (1980) Patterns of habitat partitioning by eight species of territorial caribbean damselfishes (Pisces: Pomacentridae). Bul Mar Sci 30:171–186
Weil E, Croquer A, Urreiztieta I (2009) Temporal variability and impact of coral diseases and bleaching in La Parguera, Puerto Rico from 2003–2007. Caribbean J Sci 45:221–246
Wellington GM (1992) Habitat selection and juvenile persistence control the distribution of two closely related Caribbean damselfishes. Oecologia 90:500–508. https://doi.org/10.1007/bf01875443
Wilson S, Bellwood D (1997) Cryptic dietary components of territorial damselfishes (Pomacentridae, Labroidei). Mar Ecol Prog Ser 153:299–310
Xavier R, Mazzei R, Pérez-Losada M, Rosado D, Santos JL, Veríssimo A, Soares MC (2019) A risky business? Habitat and social behavior impact skin and gut microbiomes in caribbean cleaning gobies. Front Microbiol 10:1. https://doi.org/10.3389/fmicb.2019.00716
Yan Q, Li J, Yu Y, Wang J, He Z, Van Nostrand JD, Kempher ML, Wu L, Wang Y, Liao L, Li X, Wu S, Ni J, Wang C, Zhou J (2016) Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ Microbiol 18:4739–4754. https://doi.org/10.1111/1462-2920.13365
Zeller DC (1988) Short-term effects of territoriality of a tropical damselfish and experimental exclusion of large fishes on invertebrates in algal turfs. Mar Ecol Prog Ser 44:85–93
Zemke-White LW, Beatson EL (2005) Algal community composition within territories of the damselfish Stegastes nigricans (Pomacentridae, Labroidei) in Fiji and the Cook Islands. South Pac J Nat Sci 23:43–47
Zhang Z, Li D, Xu W, Tang R, Li L (2019a) Microbiome of co-cultured fish exhibits host selection and niche differentiation at the organ scale. Front Microbiol 10:2576. https://doi.org/10.3389/fmicb.2019.02576
Zhang X, Ding L, Yu Y, Kong W, Yin Y, Zhenyu ZX, Xu Z (2019b) The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front Immunol 9:2972. https://doi.org/10.3389/fimmu.2018.02972
Acknowledgements
We thank the staff of Isla Magueyes Laboratories, and the Center for Marine and Environmental Studies at the University of the Virgin Islands for logistic support. We thank the reviewers for their suggestions that helped improve this manuscript. Field work was supported by National Science Foundation grant OCE-1536794 (PC Sikkel, PI). Laboratory work and data analysis were financed by the European Regional Development Fund (ERDF) through the COMPETE program and by National Funds through FCT—Foundation for Science and Technology (project PTDC/MAR-BIO/0902/2014 -POCI-01-0145-FEDER-016550; project PTDC/BIA-MIC/27995/2017 POCI-01-0145-FEDER-027995); RX, AP and DR were supported by FCT under the Programa Operacional Potencial Humano—Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese Ministério da Educação e Ciência (AP doctoral grant PD/BD/114357/2016; DR doctoral grant SFRH/BD/117943/2016; RX Investigador FCT contract IF/00359/2015). This is contribution #216 from the Center for Marine and Environmental Studies of the University of the Virgin Islands.
Funding
Field work was supported by National Science Foundation grant OCE-1536794 (PC Sikkel, PI). Laboratory work and data analysis were financed by the European Regional Development Fund (ERDF) through the COMPETE program and by National Funds through FCT—Foundation for Science and Technology (project PTDC/MAR-BIO/0902/2014 -POCI-01-0145-FEDER-016550; project PTDC/BIA-MIC/27995/2017 POCI-01-0145-FEDER-027995); RX, MCS, AP and DR were supported by FCT under the Programa Operacional Potencial Humano—Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese Ministério da Educação e Ciência (Andres Pagan and Ana Pereira doctoral grants PD/BD/114357/2016 and SFRH/BD/144928/2019; DR doctoral grant SFRH/BD/117943/2016; MCS DL57/2019/CP1440/CT0019; RX Investigador FCT contract IF/00359/2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical statement
The authors declare no conflict of interest and declare that this work was funded the by the European Regional Development Fund (ERDF) through the COMPETE program and by National Funds through FCT—Foundation for Science and Technology (project PTDC/MAR-BIO/0902/2014 -POCI-01-0145-FEDER-016550; project PTDC/BIA-MIC/27995/2017 POCI-01-0145-FEDER-027995; SFRH/BD/144928/2019; PD/BD/114357/2016; SFRH/BD/117943/2016; IF/00359/2015) and the National Science Foundation grant OCE-1536794 (PC Sikkel, PI). All animal subjects were handled in compliance with Arkansas State University Institutional Animal Care and Use Committee (IACUC) protocol number 778227-1, P.C. Sikkel, PI. All collecting activities were conducted as permitted by the Virgin Islands Department of Planning and Natural Resources (DPNR) and the Puerto Rico Departamento De Recursos Naturales Y Ambientales (DRNA).
Ethics approval
No human subjects were involved and, therefore, no informed consent was required. All animal subjects were handled in compliance with Arkansas State University Institutional Animal Care and Use Committee (IACUC) protocol number 778227-1, P.C. Sikkel, PI. All collecting activities were conducted as permitted by the Virgin Islands Department of Planning and Natural Resources (DPNR) and the Puerto Rico Departamento De Recursos Naturales Y Ambientales (DRNA).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Responsible Editor: Clements.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xavier, R., Pereira, A., Pagan, A. et al. The effects of environment and ontogeny on the skin microbiome of two Stegastes damselfishes (Pomacentridae) from the eastern Caribbean Sea. Mar Biol 167, 102 (2020). https://doi.org/10.1007/s00227-020-03717-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-020-03717-7