Abstract
Bdelloura candida (Platyhelminthes, Tricladida, Maricola) is an ectocommensal symbiont on the American horseshoe crab Limulus polyphemus, living on the book gills and appendages, where it spends its entire life. Given its limited dispersal capabilities and its inability to live outside of the host, we hypothesized a genetic structure that parallels that of its host. We obtained 84 planarian individuals from 19 horseshoe crabs collected from 10 sites from Massachusetts to Florida. We amplified the mitochondrial 16S rRNA and the nuclear internal transcribed spacer 2 and conducted phylogeographic and population genetic analyses, which show a clear and strong genetic break between the populations in the Atlantic and the Gulf coasts. Among the Atlantic populations, two additional, weaker barriers located along Cape Hatteras and Cape Cod restrict gene flow. Even though previous studies have suggested that the populations of the host may be in decline, those of B. candida remain stable, and some even shows signatures of expansion. Our results indicate that the phylogeography of these marine ectocommensal triclads closely mirrors that of its Limulus host, and highlight the challenges to both host and symbiont to genetically connect populations across their distribution.
Similar content being viewed by others
Introduction
The study of symbiosis is a growing field in biology, requiring integration of multiple disciplines (McFall-Ngai 2008). Symbiotic relationships among different phyla are often “loose”, especially among ectocommensal animals, which are commonly non-specific. However, many such ectocommensal relationships are known to be strictly specific, such as those of cycliophorans with their nephropid (clawed) lobsters (Funch and Kristensen 1995; Obst et al. 2006), where a faithful one-to-one species–host relationship exists, even in places where two host species coexist (Baker et al. 2007; Baker and Giribet 2007). In this case it is easy to appeal to the phenomenon of co-speciation, as each lobster host has a different ectocommensal species of Symbion, and these have never been found in non-nephropid hosts (it is thought that the American lobster can have up to three cryptic species of Symbion in the S. americanus complex, but these are not shared with any other host species). In general, symbiont and host share gene flow and their genetic structure is linked, the symbiont mirroring the genetic structure of the host (Blasco-Costa and Poulin 2013), although the global level of population genetic differentiation is usually lower in symbionts and parasites than in hosts (Mazé-Guilmo et al. 2016). However, species traits (e.g., size and life cycle) other than host genetic structure can have an effect in shaping symbiont structure, as shown in two recent meta-analyses of host-symbiont genetic structure (Blasco-Costa and Poulin 2013; Mazé-Guilmo et al. 2016).
A potentially similar case is that of the members of Bdellouridae, a family of marine planarians (Platyhelminthes, Tricladida, Maricola) and its host, the American horseshoe crab, Limulus polyphemus (Linnaeus, 1758). The members of the genus Bdelloura are characterized by the absence of eye lenses, a posterior adhesive caudal disk set off from the rest of the body, and numerous testes distributed throughout the body (Leidy 1851). The genus includes three described species, Bdelloura candida (Girard, 1850), B. propinqua Wheeler, 1894, and B. wheeleri Wilhelmi, 1909, of which B. candida is the most widespread and easily identified (Sluys 1989).
Bdelloura candida was described from Chelsea Beach, Massachusetts (Girard 1850). Their whitish-coloured, oval-shaped bodies are about 15 × 4 mm (sometimes up to 2 cm in length) while moving, according to Wilhelmi (1908). In his monograph, Sluys (1989) described B. candida’s characteristic undulated sides, large central pharynx, and broad caudal disk that changes shape depending on the state of contraction. Bdelloura candida lives ectocommensally on the walking appendages, carapace, and book gills of the Atlantic horseshoe crab. Despite relying heavily on their hosts for indirect nutrition, the feeding mechanisms and digestive structures of the commensal species of Bdellouridae do not differ significantly from those of their free-living relatives (Jennings 1977). Bdelloura candida can be found concomitantly with its host throughout its distribution range along the Gulf and Atlantic coasts of North America (Sekiguchi and Shuster 2009). As a direct developer (Sluys 1989), B. candida hatches from cocoons attached to the book gills of L. polyphemus, and it has, therefore, little capacity for dispersal. The adults cannot survive independently, living exclusively on its L. polyphemus host; thus, they must recolonize the same host or a nearby individual after Limulus moults, a process that probably relies on chemical signalling (Chevalier and Steinbach 1969). This strict association to horseshoe crabs makes B. candida an excellent model for phylogeographic research of symbiotic organisms.
The Atlantic horseshoe crab is distributed along the east coast of North America, from Maine to Florida, with additional populations in the eastern Gulf of Mexico and around the Yucatan peninsula (Anderson and Shuster 2003). Along this range, distinct populations of L. polyphemus are recognized (Shuster 1982; King et al. 2015), and their population sizes show a clear latitudinal gradient, the largest densities being found towards the middle of the range, in and surrounding Delaware Bay (e.g., Botton and Ropes 1987; Shuster 2003). However, many of these populations have suffered recent decline due to a diversity of anthropogenic factors (Faurby et al. 2010). Genetically, a marked structure exists among specimens north and south of northeastern Florida in mitochondrial DNA (Saunders et al. 1986a), allozymes (Selander et al. 1970), microsatellite nuclear markers (King et al. 2015), and morphology (Riska 1981), in what constitutes a well-documented marine biogeographic break (e.g., Avise 1992; Lee and Foighil 2004). This genetic structure most probably results from the territoriality of adults and the low dispersal capacity of the trilobite larva, which has been mainly studied in the Delaware estuary (Botton and Loveland 1987). Their transport as passive particles by the alongshore currents was estimated between 144 and 432 m per h, although they tend to remain in the close vicinity of the shoreline (Botton and Loveland 2003). Recapture data from L. polyphemus tag-and-release experiments provide a mean recovery distance of approximately 3–4 miles (Baptiste et al. 1957; Sokoloff 1978). Furthermore, many adults were shown to remain tightly associated with their particular estuary or local shoreline. This philopatry regarding reproductive and behavioural habits is also suggestive of a pattern of genetic structure.
Several authors have already postulated that genetic structure from some symbionts/parasites can mirror and even complement genetic data from their hosts (Nieberding and Olivieri 2007). Since genetic data are not available for B. candida to test whether its genetic structure mirrors that of L. polyphemus, we examined specimens from hosts at ten sites along the Atlantic and Gulf coasts of the USA, as we wanted to ascertain whether the commensal showed the signature of the host’s phylogeographic structure.
Materials and methods
Sample collection
A total of 84 specimens of B. candida were collected from 18 L. polyphemus individuals at ten sampling sites along the Atlantic and Gulf coasts (Table 1). Specimens were fixed in 96% ethanol and stored at −80 °C for long-term preservation. All host and planarian specimens are deposited in the Invertebrate Zoology collections in the Museum of Comparative Zoology (MCZ), Harvard University.
Molecular analyses
DNA was extracted using the DNEasy® tissue kit (Qiagen, Valencia, CA, USA) following manufacturer’s instructions, with a slight modification in the incubation of the tissue in lysis buffer (overnight). We selected two markers, the mitochondrial 16S rRNA and the nuclear ribosomal internal transcribed spacer-2 region (ITS2). The 16S rRNA marker was amplified in 83 individuals with the primer pair 16Sa–16Sb (16Sa: 5′-CGC CTG TTT ATC AAA AAC AT-3′, 16Sb; 5′-CTC CGG TTT GAA CTC AGA TCA-3′; Xiong and Kocher 1991) to target a region of approximately 395 base pairs. The ITS2 marker was amplified in 52 individuals using universal primers 5.8SF (Carranza 1997) and 28S rev (Lôbo-Hajdu et al. 2003–2004; Duran et al. 2004). It is important to note here that we failed to sequence ITS2 from all individuals, in part because of an apparent indel-level variation within the many copies of the rRNA cistron, which may prevent correct amplification and/or direct Sanger sequencing of PCR products.
PCR reactions were conducted using 1 μL of template DNA, 18 μL molecular grade distilled water, 5 μL GoTaq ® Buffer (Promega, Madison, WI, USA), 0.5 μL 10 mM dNTPs (New England Biolabs, Inc., Ipswich, MA, USA), 0.13 μL GoTaq® Taq Polymerase (Promega, Madison, WI, USA), and 0.25 μL of each forward and reverse primers (10 mM). The PCR program for both markers included an initial denaturing step at 95 °C for 2 min, 35 cycles of denaturing at 95 °C for 30 s, annealing at 45 (for 16S rRNA) to 49 °C (for ITS2) for 30 s, and extension at 72 °C for 1 min, and then a final extension step at 72 °C for 1 min. The PCR products were vacuum cleaned and sequencing reactions were done using BigDye Terminator v3.1 (Life Technologies, Carlsbad, CA, USA) and a sequencing program with an initial denaturing step at 94 °C for 3 min, then 25 cycles of denaturing at 94 °C for 10 s, annealing at 50 °C for 5 s, and extension at 60 °C for 4 min. The purified PCR products were sequenced using the same primer pairs in a 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA) at the Harvard Center for Systems Biology.
Chromatograms were edited and assembled using Sequencher 5.0.1 (Gene Codes Corporation 1991–2011, Ann Arbor, MI, USA). Sequences were checked for contamination using BLAST (Altschul et al. 1997) on the NCBI website (http://ncbi.nlm.nih.gov). Accession numbers for 16S sequences in GenBank are KY499968-KY500051 (84 sequences) and those of ITS2 are KY500082-KY500133 (52 sequences).
Sequence alignment
Sequence data were aligned using MAFFT version 7 (Katoh et al. 2002; Katoh and Standley 2013) on the developers’ online server (http://mafft.cbrc.jp/alignment/server/). For 16S rRNA, the L-INS-i strategy was used (Katoh et al. 2005). Because the DNA sequences were closely related, the scoring matrix for nucleotide sequences was set to 1PAM/k = 2. The offset value was set to 0.1 and the gap opening penalty (for all molecular markers) was 1.53. ITS-2 had similar settings, with the only change being the E-INS-i strategy (very slow, for sequences with multiple conserved domains and large gaps) (Katoh et al. 2005). Final length of the alignments after trimming the leading and trailing ends was 346 bp for 16S rRNA and 1248 bp for ITS2.
Genetic diversity indices
Number of haplotypes and segregating sites, haplotype diversity, nucleotide diversity and mean number of pairwise differences were obtained in DNAsp 5.10.1 (Librado and Rozas 2009). Haplotype networks were estimated with TCS (Clement et al. 2000) implemented in PopART (http://popart.otago.ac.nz).
Distribution of the genetic diversity
Genetic differentiation between populations was measured by computing pairwise Ф ST statistics in ARLEQUIN v 3.5 (Excoffier and Lischer 2010), evaluating the corresponding p values by 10,000 permutations, which were then adjusted using a false discovery rate (Narum 2006). To test whether the data followed a pattern of isolation by distance, we performed a Mantel test in Genepop (Raymond and Rousset 1995) with 16,000 permutations, and converting the Ф ST matrix to Ф ST/1 − Ф ST. In this case, only the 16S rRNA dataset, which contained all populations, was analysed. Geographical distances were calculated as the minimum distance in kilometres between sites. The effect of barriers in determining the genetic structure of B. candida populations was further evaluated using pairwise Ф ST values and visualized with the software BARRIER v2.2 (Manni et al. 2004). BARRIER uses the matrix of geographical coordinates and links it with the corresponding distance matrix (Ф ST), and subsequently applies Monmonier’s distance algorithm to identify areas that hinder gene flow among sites, i.e., the zones where Ф ST differences between pairs of sites are the largest.
Differentiation between groups of populations was assessed by conducting hierarchical analyses of molecular variance (AMOVA) using genetic distances, and testing their significance by running 16,000 permutations in ARLEQUIN. We grouped the populations in the following sets, based on results from BARRIER (see “Results” section): Gulf (FL1, FL2, FL3), Atlantic (GA, SC, NC, DE, CT, MA1, MA2), Group 1 (GA, SC, NC, DE, CT), Group 2 (GA, SC, NC), Group 3 (DE, CT), and Group 4 (MA1, MA2). We quantified three components of the genetic variance as follows: among groups [using three different combinations: (a) Gulf vs. Atlantic, (b) Gulf vs. Group 1 vs. Group 2, and (c) Gulf vs. Group 3 vs. Group 4], among populations within groups, and within populations. AMOVA analyses were conducted on the 16S rRNA and the ITS2 datasets separately.
Demographic analyses
We performed the Tajima’s D test of neutrality to test for bottlenecks and population expansions (Tajima 1989). The history of effective population size was assessed through the pairwise mismatch distribution in ARLEQUIN, where populations in expansion show unimodal distributions while stationary populations show multimodal distributions. In addition, the neutrality Ramos-Onsins and Rozas’ R2 test was calculated for the four groups of populations obtained from Barrier [Gulf (FL1, FL2, FL3), Group 2 (GA, SC, NC), Group 3 (DE, CT), and Group 4 (MA1, MA2)] (Ramos-Onsins and Rozas 2002).
Given the high genetic divergence detected between the populations of Florida (“Gulf”) and those from the rest of the Eastern coast of the US (“Atlantic”) shown in “Results” section, we conducted a Bayesian analysis to assess migration rates between both regions in LAMARC v 2.1.9 (Kuhner 2006). Two initial runs were conducted to estimate the most likely priors for our dataset. Then, we used the Felsenstein’s 84 (F84) model of DNA sequence evolution and variation values of migration between 0 and 30 in both directions. The final run was based on four different replicates with 10 initial MCMC chains of 5000 generations each, a burning period of 500, followed by 2 final MCMC chains of 40,000 generations each with a burning period of 1000. Two simultaneous heating searches (1 and 1.5) were performed per replicate. To test convergence of the chains and confirm the existence of at least 200 independent simulations (effective sample size—ESS) for each parameter, results were summarised in Tracer v 1.5 (http://beast.bio.ed.ac.uk/Tracer). Migration rates (Mt) were expressed as the number of migrants per generation Mt = n/µ, where n is the immigration rate per generation, and µ the substitution rate.
Results
Genetic diversity and variation in Bdelloura candida
All values of genetic diversity are detailed in Table 2. Surprisingly, for most populations, haplotype diversity was higher for the ITS2 than for 16S rRNA, given that less individuals were sequenced for ITS2 than for 16S rRNA. We found 11 private haplotypes for each marker (68.75% of total haplotypes for 16S rRNA and 55% for ITS2). Values of nucleotide diversity were generally low (<0.0055) except for FL3 and NC (for the 16S rRNA dataset). The haplotype networks of both markers (Fig. 1) showed a clear separation between the populations along the Gulf of Mexico (FL1, FL2, and FL3) and the haplotypes found in the populations of the East Atlantic coast (MA1, MA2, CT, DE, NC, SC, and GA), connected by 15 mutational steps in the 16S rRNA network and three in ITS2 (Fig. 1). There were no shared haplotypes between the populations of the Gulf of Mexico and those of the Atlantic coast (Fig. 1). The 16S rRNA haplotype network contained a single common haplotype distributed in all populations of the Atlantic coast and six private haplotypes (Fig. 1), while in the Gulf coast, we found one haplotype distributed in ten individuals, three more haplotypes in three individuals, and five private haplotypes (Fig. 1). The ITS2 haplotype network showed two main haplotypes in the Atlantic coast without a clear geographic pattern and seven private haplotypes (Fig. 1), while in the Gulf we found one main haplotype and three private haplotypes (Fig. 1).
When the populations were analysed separately to evaluate the genetic diversity contained per host, we observed that not all individuals of B. candida collected from the same host were genetically identical (Fig. 2). In turn, 2–6 haplotypes were found within the same horseshoe crab host for both markers (Figs. 2, 3). When several hosts were collected, in some cases only one haplotype was recovered for B. candida in all hosts, for instance in Connecticut and Delaware for 16S rRNA (Fig. 3), while in most cases more than one Bdelloura haplotypes were collected in each host, especially for ITS2 (Fig. 3).
The Ф ST values for both markers showed mainly significant differences between pairwise comparisons among populations from the Gulf of Mexico and those from the Atlantic coast (Table 3). We found that both the Atlantic coast and the Gulf populations were panmictic (Table 3).
Using the matrix of Ф ST values for 16S rRNA and the geographic coordinates, BARRIER located the three a priori selected barriers in Cape Canaveral, Cape Hatteras and Cape Cod (Fig. 4a, b), grouping the populations in four groups (Gulf, Groups 1, 3, and 4). All AMOVA analyses for both markers showed significant differences among groups and among individuals within populations (Tables 4, 5). However, the percentage of variation obtained when analysing the differences among groups was highest when considering four groups (Gulf vs. Group 1 vs. Group 3 vs. Group 4) for 16S rRNA (Table 4) and when analysing three groups (Gulf vs. Group 1 vs. Group 2) for ITS2 (Table 5). This genetic differentiation could be attributed to a pattern of isolation by distance (Supplementary Figure 1; p = 0.005 for 16S rRNA and p = 0.0105 for ITS2).
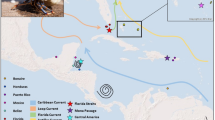
Location of major genetic breaks and oceanographic fronts in Bdelloura candida. A Geographic location of three major genetic breaks in the 16S rRNA dataset ranked in order of importance from a to c. Polygons indicate Delaunay triangulations obtained from Voronoï tessellations. B Reconstruction of seasonal averages for major currents using Mariano Global Surface Velocity Analysis (MGSVA) in the North Atlantic coast of the United States (from Gyory, J. Mariano, A.J., Ryan, E.H. The Gulf Stream. Ocean Surface Currents. http://oceancurrents.rsmas.miami.edu/atlantic/gulf-stream.html)
Demographic analyses
Neutrality tests were conducted for those populations for which more than three individuals were available. The Tajima’s D statistics were not significant in any of the populations, indicating no evidence of selection (Table 2), and the mismatch distribution plots suggested population expansions in almost all populations analysed (Supplementary Figure 2). When grouping the populations into four groups (Gulf, Group 2, Group 3 and Group 4), the Ramos-Onsins & Rozas’ R2 test was significant for the Gulf populations for both markers and also for Group 3 (DE, CT) for ITS2 (Table 6), and therefore, a null hypothesis of neutral evolution under a constant population size can be rejected. However, Tajima’s D was significant only in Group 2 (GA, SC, NC) for the 16S rRNA marker (Table 6). LAMARC did not show clear patterns of migration in any direction (Fig. 5) between the two main groups of populations (Gulf to Atlantic or Atlantic to Gulf).
Graphical representation of the results for the migration analysis between Atlantic and Gulf populations in Bdelloura candida using the 16S rRNA and ITS2 markers. The four graphs show the five replicates used in LAMARC to analyse the migration rate (Mt) from the Gulf to the Atlantic (left graphs) and from the Atlantic to the Gulf of Mexico (right graphs)
Discussion
Genetic diversity estimates for Bdelloura candida
The low values of nucleotide diversity (π) of B. candida were similar to those of other triclads (e.g., Sunnucks et al. 2006; Álvarez-Presas et al. 2012), although these are terrestrial free-living species, which makes the comparisons less relevant given the peculiarities of their habitats. The Ramos-Onsins & Rozas’ R2 and Tajima’s D neutrality tests were mostly non-significant, which indicates a certain level of stability, although sample sizes were small, and therefore, conclusions from Tajima’s D test should be taken with caution. However, the Gulf region did show evidence of population expansion for both markers, as well as the group comprised by North Carolina, South Carolina and Georgia for 16S rRNA when using Ramos-Onsins & Rozas R2 test, which is robust to small sample sizes. Signatures of population expansion were also detected in the group comprised by Delaware and Connecticut for ITS2. However, such values could also be the consequence of purifying selection acting on the mitochondrial or nuclear genomes. Even if populations of B. candida were not in expansion but remained stable, these results are intriguing since the host L. polyphemus has been in decline over the recent decades (Gómez-Aguirre 1993; Faurby et al. 2010), and one could expect a similar trend in the viability of the populations of both the symbiont and the host. Perhaps, since each host can maintain up to dozens of bdellourid specimens, a decline in the horseshoe crabs should actually reach a certain threshold before affecting the genetic diversity in the symbionts.
Given the life cycle of B. candida, where adults have to change host during or after moulting, and therefore, a random pool of haplotypes might be colonizing the horseshoe crabs each time, low levels of clonality were expected. Indeed, in some cases we found high haplotypic diversity within the same individual host, with 2–6 haplotypes appearing concomitantly for both markers.
Phylogeographic patterns: genetic breaks and geographic barriers
The genetic structure of B. candida along the Atlantic coast of the United States mirrors that of its host, L. polyphemus, as usually occurs in host–symbiont relationships (Mazé-Guilmo et al. 2016). In our results, the correlation of geographic distance and genetic differentiation in both genetic markers indicates a pattern of isolation by distance, also recognized in L. polyphemus populations (King et al. 2015), which is compatible with certain level of genetic structuring along the Atlantic coast of North America. The results of the haplotype networks, Ф ST, and AMOVA analyses indicate a significant genetic break between the Atlantic and Gulf coast populations of B. candida, which the software BARRIER locates primarily along the Atlantic coast of Florida. Geographic and ecological barriers may influence the dispersal capabilities of an organism, as well as its biological features limiting potential for long-distance dispersal and playing a role in genetic and geographic variation of a species (Gawarkiewicz et al. 2007). In this sense, it seems that the Florida peninsula represents a strong barrier to the already-limited dispersal capacity of B. candida.
Several authors have also previously found a sharp genetic break within the continuously distributed Atlantic and Gulf coast populations of L. polyphemus (Selander et al. 1970; Riska 1981; Saunders et al. 1986b; King et al. 2015), which parallels our analyses of B. candida. Riska (1981) found that populations of L. polyphemus from Cape Cod to Mexico showed unusually large morphological differences, exhibiting a north–south cline associated with the location of particular spines on the body and the length of the doublure anterior to the mesial spine. The major genetic break for L. polyphemus occurs in Northern Florida, approximately in the Indian River region, a region long-recognized as a transition zone between temperate and tropical marine ecosystems and fauna that exhibit analogous phylogeographic patterns (Briggs 1974). The Indian River region is known for its peculiarities, showing highly variable salinity measurements depending on distance from the inlets and the magnitude of freshwater inputs (Ehlinger and Tankersley 2004), which indeed hampers gene flow in L. polyphemus (King et al. 2015) and potentially in its symbionts.
Another physical barrier located in Florida, potentially responsible for the genetic structure of B. candida, is surface temperature and movement of water masses, since the limits of the L. polyphemus distribution are mostly determined by temperature (Sekiguchi and Shuster 2009). The temperature clines of approximately 24–27 and 27–30 °C surrounding southern Florida are visible in both day and night observations, and may represent a significant water mass boundary that isolates the Atlantic Ocean and Gulf of Mexico populations of ectocommensal marine triclads. In addition to physical barriers, ecological barriers in this region may also help explain the genetic break between the Atlantic and Gulf populations. The rapid urbanization of the Atlantic coast of Florida has severely impacted the beaches (Halpern et al. 2008), destroying the available habitats for L. polyphemus (Walls et al. 2002; Shuster 2003; Shuster and Sekiguchi 2009; Faurby et al. 2010).
The divergence in the genetic structure of the populations of B. candida from the Gulf of Mexico and the Atlantic coast is also maintained due to the scarce migration detected among the regions in both directions, even though the Shelf break frontal jet and the Gulf Stream may sustain some level of gene flow in this area for other organisms (Hare and Avise 1998; Gawarkiewicz et al. 2007). The Gulf Stream allows genetic connectivity across major barriers for some species (Gawarkiewicz et al. 2007), although in some instances it is insufficient and leads to lineage diversification (Boehm et al. 2013). Given that B. candida cannot survive outside its host for long periods and lacks a larval phase, gene flow between divergent regions is presumably only allowed when the adult host migrates. In this sense, the strong territoriality of the horseshoe crabs (Botton and Loveland 2003), together with the well-established oceanographic and physical barriers, severely restrict the genetic connectivity of B. candida.
Two additional barriers to gene flow in B. candida were also located along the US East coast between North Carolina and Virginia and between Rhode Island and Massachusetts (Fig. 2). These two barriers correspond well with the fronts created by the Gulf Stream flowing to Cape Hatteras, and Cape Cod isolating the colder waters of the Gulf of Maine. Indeed, around Cape Hatteras, there is a strong eastward flow of the Gulf Stream, which could constitute an effective barrier to gene flow (Fig. 2b). In addition, time/averaged flow plots of surface ocean currents (Fig. 4b) shows recirculation of waters along the coasts between Virginia and Massachusetts (also seen in Maximenko et al. 2009). Such fronts have shown strong influence on the pattern of genetic differentiation between the populations of the Atlantic coast of both B. candida and L. polyphemus (Selander et al. 1970; Saunders et al. 1986a; King et al. 2015), as well as in other marine organisms (see reviews by Avise 1992, 1996). However, the genetic structure of B. candida is not entirely mirrored by that of L. polyphemus, which showed great similarities in the genetic patterns of horseshoe crab populations of Connecticut, Delaware and North Carolina (King et al. 2015), while in B. candida North Carolina groups with South Carolina and Georgia (Fig. 4). Denser sampling along this region may serve to refine these barriers.
Climate-related factors influencing genetic structure in Bdelloura candida
Bdelloura candida’s patterns of genetic variation can be influenced by other factors besides present-day physical and oceanographic barriers, among which climate may be one of the strongest factors. It is likely that the conditions of the last glacial maximum dramatically affected L. polyphemus population sizes and that these populations gradually expanded their range northward when the ice receded, but their genetic diversity had been severely compromised (Faurby et al. 2010), at least in the northern range of its distribution. Genetic signatures of a recent post-glacial population expansion in B. candida can be hinted in the 16S rRNA haplotype network (Fig. 1), with a characteristic star-like structure showing private haplotypes radiating from a single dominant haplotype, although the small sampling size makes this conclusion tentative.
In addition to the end of the last ice age, major climactic oscillations have contributed to shifting ranges of species in the North Atlantic (e.g., Maggs et al. 2008). These changes in climate affect the genetic structure and diversity of populations, and even entire species (Faurby et al. 2010). These post-Pleistocene climate fluctuations have also influenced physical and ecological properties of the east coast of North America, potentially causing both ancient divergence and some of the geographic barriers that still exist today, e.g., present-day coastal currents and ocean circulation patterns, and glacial moraines like Long Island and Cape Cod (Faurby et al. 2010).
Conclusion
There is a strong agreement in all our molecular analyses indicating a major genetic break between the Gulf and Atlantic coast populations of B. candida, a break that has been identified in many other marine organisms (Avise 1992). In this sense, the phylogeography of these marine parasitic triclads closely mirrors that of its host. However, future studies examining the genetic diversity and geographic differentiation of B. candida, or marine triclads in general, are needed to elucidate the exact location of the boundary zone between the Atlantic and Gulf coast populations. Southeastern Florida, in particular, is poorly sampled for both L. polyphemus and B. candida, as animals are much more difficult to find in the southern ranges of their distribution. In this context, future sampling in the disjunct populations known from the Yucatan peninsula should also help to refine southerly geographic barriers in this species. In addition, given the strong nature of the major barrier isolating the populations from the Gulf of Mexico from those of the Atlantic coast of the United States, morphological analyses of B. candida are needed to help elucidate potential speciation processes that are also currently hinted in the host.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Álvarez-Presas M, Mateos E, Vila-Farré M, Sluys R (2012) Riutort M (2012) Evidence for the persistence of the land planarian species Microplana terrestris (Muller, 1774) (Platyhelminthes, Tricladida) in microrefugia during the Last Glacial Maximum in the northern section of the Iberian Peninsula. Mol Phylogenet Evol 64:491–499. doi:10.1016/J.Ympev.2012.05.001
Anderson LI, Shuster CN Jr (2003) Throughout geologic time: where have they lived? In: Shuster CN Jr, Barlow RB, Brockmann HJ (eds) The American horseshoe crab. Harvard University Press, Cambridge, pp 189–223
Avise JC (1992) Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos 63:62–76
Avise JC (1996) Toward a regional conservation genetics perspective: phylogeography of faunas in the southeastern United States. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman & Hall, New York, pp 431–470
Baker JM, Giribet G (2007) A molecular phylogenetic approach to the phylum Cycliophora provides further evidence for cryptic speciation in Symbion americanus. Zool Scr 36:353–359
Baker JM, Funch P, Giribet G (2007) Cryptic speciation in the recently discovered American cycliophoran Symbion americanus; genetic structure and population expansion. Mar Biol 151:2183–2193
Baptiste JP, Smith OR, Ropes JW (1957) Migration of the horseshoe crab Limulus polyphemus in Plum Island Sound, Massachusetts. Special Sci Rep Fish 220:1–15
Blasco-Costa I, Poulin R (2013) Host traits explain the genetic structure of parasites: a meta-analysis. Parasitology 140:1316–1322. doi:10.1017/S0031182013000784
Boehm JT, Woodall L, Teske PR, Lourie SA, Baldwin C, Waldman J, Hickerson M (2013) Marine dispersal and barriers drive Atlantic seahorse diversification. J Biogeogr 40:1839–1849. doi:10.1111/jbi.12127
Botton ML, Loveland RE (1987) Orientation of the horseshoe crab, Limulus polyphemus, on a sandy beach. Biol Bull 173:289–298. doi:10.2307/1541542
Botton ML, Loveland RE (2003) Abundance and dispersal potential of horseshoe crab (Limulus polyphemus) larvae in the Delaware estuary. Estuaries 26:1472–1479. doi:10.1007/Bf02803655
Botton ML, Ropes JW (1987) Populations of horseshoe crabs, Limulus polyphemus, on the Northwestern Atlantic continental shelf. Fish Bull 85:805–812
Briggs JC (1974) Marine zoogeography. McGraw-Hill Book Company, London
Carranza S (1997) Taxonomia molecular mitjançant la seqüenciació del DNA ribosòmic 18S: Aplicació a l’origen i filogènia dels Platihelmints. Ph.D. Departament de Genètica, Barcelona
Chevalier RL, Steinbach HB (1969) A chemical signal attracting the flatworm Bdelloura candida to its host, Limulus polyphemus. Biol Bull 137:394
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Duran S, Giribet G, Turon X (2004) Phylogeographic history of the sponge Crambe crambe (Porifera, Poecilosclerida): range expansion and recent invasion of Macaronesian islands from the Mediterranean Sea. Mol Ecol 13:109–122
Ehlinger GS, Tankersley RA (2004) Survival and development of horseshoe crab (Limulus polyphemus) embryos and larvae in hypersaline conditions. Biol Bull 206:87–94. doi:10.2307/1543539
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Faurby S, King TL, Obst M, Hallerman EM, Pertoldi C, Funch P (2010) Population dynamics of American horseshoe crabs-historic climatic events and recent anthropogenic pressures. Mol Ecol 19:3088–3100
Funch P, Kristensen RM (1995) Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature 378:711–714
Gawarkiewicz G, Monismith S, Largier J (2007) Observing larval transport processes affecting population connectivity progress and challenges. Oceanography 20:40–53
Girard C (1850) Several new species of marine Planariae of the coast of Massachusetts. Proc Boston Soc Nat History 3:251
Gómez-Aguirre S (1993) Cacerolita de mar (Limulus polyphemus L.) en la Península de Yucatán Biodiversidad Marina y Costera de México. CONABIO & CIQRO, Mexico, pp 650–659
Halpern BS, Walbridge S, Selkoe KA, Kappel CV, Micheli F, D’Agrosa C, Bruno JF, Casey KS, Ebert C, Fox HE, Fujita R, Heinemann D, Lenihan HS, Madin EMP, Perry MT, Selig ER, Spalding M, Steneck R, Watson R (2008) A global map of human impact on marine ecosystems. Science 319:948–952. doi:10.1126/Science.1149345
Hare MP, Avise JC (1998) Population structure in the American oyster as inferred by nuclear gene genealogies. Mol Biol Evol 15:119–128
Jennings JB (1977) Patterns of nutritional physiology in free-living and symbiotic Turbellaria and their implications for the evolution of entoparasitism in the phylum Platyhelminthes. Acta Zool Fennica 154:63–79
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/Molbev/Mst010
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi:10.1093/nar/gki198
King TL, Eackles MS, Aunins AW, Brockmann HJ, Hallerman E, Brown BL (2015) Conservation genetics of the American horseshoe crab (Limulus polyphemus): Allelic diversity, zones of genetic discontinuity, and regional differentiation. In: Carmichael RH (ed) Changing global perspectives on horseshoe crab biology, conservation and management. Springer International Publishing, Cham, pp 65–96
Kuhner MK (2006) Robustness of coalescent estimators to between-lineage mutation rate variation. Mol Biol Evol 23:2355–2360. doi:10.1093/molbev/msl106
Lee T, Foighil DÓ (2004) Hidden Floridian biodiversity: mitochondrial and nuclear gene trees reveal four cryptic species within the scorched mussel, Brachidontes exustus, species complex. Mol Ecol 13:3527–3542
Leidy J (1851) Helminthological contributions—No. 3. Proc of the Acad Nat Sci Phila 5:239–254
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lôbo-Hajdu G, Guimarães ACR, Salgado A, Lamarão FRM, Vieiralves T, Mansure JJ, Albano RM (2003–2004) Intragenomic, intra- and interspecific variation in the rDNA ITS of Porifera revealed by PCR-single-strand conformation polymorphism (PCR-SSCP). Bollettino dei Musei e degli Istituti Biologici dell’Università di Genova 68:413–423
Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Vainola R, Viard F, Wares J (2008) Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89:S108–S122
Manni F, Guérard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol 76:173–190
Maximenko N, Niiler P, Centurioni L, Rio M-H, Melnichenko O, Chambers D, Zlotnicki V, Galperin B (2009) Mean dynamic topography of the ocean derived from satellite and drifting buoy data using three different techniques. J Atmos Ocean Technol 26:1910–1919. doi:10.1175/2009jtecho672.1
Mazé-Guilmo E, Blanchet S, McCoy KD, Loot G (2016) Host dispersal as the driver of parasite genetic structure: a paradigm lost? Ecol Lett 19:336–347
McFall-Ngai M (2008) Are biologists in ‘future shock’? Symbiosis integrates biology across domains. Nat Rev Microbiol 6:789–792. doi:10.1038/Nrmicro1982
Narum SR (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv Genet 7:783–787. doi:10.1007/s10592-005-9056-y
Nieberding CM, Olivieri I (2007) Parasites: proxies for host genealogy and ecology? Trends Ecol Evol 22:156–165. doi:10.1016/j.tree.2006.11.012
Obst M, Funch P, Kristensen RM (2006) A new species of Cycliophora from the mouthparts of the American lobster, Homarus americanus (Nephropidae, Decapoda). Org Divers Evol 6:83–97
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Riska B (1981) Morphological variation in the horseshoe crab Limulus polyphemus. Evolution 35:647–658
Saunders NC, Kessler LG, Avise JC (1986) Genetic variation and geographic differentiation in mitochondrial DNA of the horseshoe crab, Limulus polyphemus. Genetics 112:613–627
Sekiguchi K, Shuster CN (2009) Limits on the global distribution of horseshoe crabs (Limulacea): lessons learned from two lifetimes of observations: Asia and America. Biol Conserv Horseshoe Crabs. doi:10.1007/978-0-387-89959-6_1
Selander RK, Yang SY, Lewontin RC, Johnson WE (1970) Genetic variation in the horseshoe crab (Limulus polyphemus), a phylogenetic “relic”. Evolution 24:402–414
Shuster CN Jr (1982) A pictorial review of the natural history and ecology of the horseshoe crab, Limulus polyphemus, with reference to other Limulidae. In: Bonaventura J, Bonaventura C, Tesh S (eds) Physiology and biology of horseshoe crabs: studies on normal and environmentally stressed animals. Alan R. Liss Inc, New York, pp 1–52
Shuster CN Jr (2003) King crab fertilizer: a once-thriving Delaware Bay industry. In: Shuster CN Jr, Barlow RB, Brockmann HJ (eds) The American horseshoe crab. Harvard University Press, Cambridge, pp 341–357
Shuster CN, Sekiguchi K (2009) Basic habitat requirements of the extant species of horseshoe crabs (Limulacea). Biol Conserv Horseshoe Crabs. doi:10.1007/978-0-387-89959-6_7
Sluys R (1989) A monograph of the marine triclads. A.A. Balkema, Rotterdam
Sokoloff A (1978) Observations on populations of the horseshoe crab Limulus (=Xiphosura) polyphemus. Res Popul Ecol (Kyoto) 19:222–236
Sunnucks P, Blacket MJ, Taylor JM, Sands CJ, Ciavaglia SA, Garrick RC, Tait NN, Rowell DM, Pavlova A (2006) A tale of two flatties: different responses of two terrestrial flatworms to past environmental climatic fluctuations at Tallaganda in montane southeastern Australia. Mol Ecol 15:4513–4531. doi:10.1111/j.1365-294X.2006.03107.x
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Walls EA, Berkson J, Smith SA (2002) The horseshoe crab, Limulus polyphemus: 200 million years of existence, 100 years of study. Rev Fish Sci 10:39–73. doi:10.1080/20026491051677
Wilhelmi J (1908) On the North American marine triclads. Biol Bull 15:1–6
Xiong B, Kocher TD (1991) Comparison of mitochondrial DNA sequences of seven morphospecies of black flies (Diptera: Simuliidae). Genome 34:306–311
Acknowledgements
This research, which constituted the basis for a senior thesis of E.A.B., was funded by two Museum of Comparative Zoology’s Grant for Undergraduate Research and by the Harvard Program for Research in Science and Engineering. Will Burke assisted in the field, driving and collecting specimens on the 4000+ mi collection trip in May 2013. For their hospitality during fieldwork, S. Dudley, A. Griffin, L. DiNicola, S. Farquhar, D. Wiley, and L. Nelson are specially acknowledged. In addition, Kevin Kocot kindly provided samples from Florida. The laboratory work was supported by internal funds from the MCZ. This manuscript was written during a sabbatical of G.G. in the laboratory of A.R. at The Natural History Museum, London.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest. The work was done under all the necessary permits and standards. Horseshoe crabs were released after their ectocommensals were collected, and there are no ethic protocols for collecting flatworms.
Additional information
Responsible Editor: T. Reusch.
Reviewed by A. Waeschenbach and an undisclosed expert.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Riesgo, A., Burke, E.A., Laumer, C. et al. Genetic variation and geographic differentiation in the marine triclad Bdelloura candida (Platyhelminthes, Tricladida, Maricola), ectocommensal on the American horseshoe crab Limulus polyphemus . Mar Biol 164, 111 (2017). https://doi.org/10.1007/s00227-017-3132-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3132-y