Abstract
The emergence of ocean acidification as a significant threat to calcifying organisms in marine ecosystems creates a pressing need to understand the physiological and molecular mechanisms by which calcification is affected by environmental parameters. We report here, for the first time, changes in gene expression induced by variations in pH/pCO2 in the widespread and abundant coccolithophore Emiliania huxleyi. Batch cultures were subjected to increased partial pressure of CO2 (pCO2; i.e. decreased pH), and the changes in expression of four functional gene classes directly or indirectly related to calcification were investigated. Increased pCO2 did not affect the calcification rate and only carbonic anhydrase transcripts exhibited a significant down-regulation. Our observation that elevated pCO2 induces only limited changes in the transcription of several transporters of calcium and bicarbonate gives new significant elements to understand cellular mechanisms underlying the early response of E. huxleyi to CO2-driven ocean acidification.
Similar content being viewed by others
Introduction
The oceans are the largest active sinks of carbon on Earth, with an estimated 30% of anthropogenic carbon emissions produced since 1800 taken up by oceans (Sabine et al. 2004). This leads to profound changes in the carbonate chemistry of seawater with an increase in pCO2, dissolved inorganic carbon (DIC) and bicarbonate ions (HCO3 −) concentration, and a decrease in the concentration of carbonate ions (CO3 2−) and pH. These changes are collectively referred to as ocean acidification, an anthropogenic perturbation that has been identified as a great threat to marine ecosystems (Halpern et al. 2008) and particularly to calcifying organisms (Orr et al. 2005). A decreased availability of carbonate ions could thus affect the ability of calcifying organisms to precipitate CaCO3. This will directly impact marine ecosystems by weakening CaCO3 skeletons and it will impact the ocean carbon pump as CaCO3 is thought to enhance the export of organic carbon in the deep ocean (“carbon ballasting”; Engel et al. 2009). Coccolithophores are the dominant planktonic calcifiers in the present ocean and are estimated to be responsible for about half of all modern precipitation of CaCO3 (Milliman 1993). Thus it is crucial to understand how these organisms will be affected by ocean acidification in order to effectively predict the response of the ocean to this large-scale perturbation and its future ability to absorb anthropogenic CO2.
A large range of coccolithophores responses to elevated pCO2 have been observed in laboratory cultures (Riebesell et al. 2000; Zondervan et al. 2001; Langer et al. 2006, 2009; Iglesias-Rodriguez et al. 2008; Ridgwell et al. 2009; Shi et al. 2009; Müller et al. 2010). Resolving this diversity in responses requires a better understanding of the cellular and biochemical mechanisms and pathways involved in calcification and how they are affected by changes in pCO2 and other environmental parameters. The molecular mechanisms involved in coccolithophore biomineralization are still poorly understood despite extensive physiological investigation (reviewed by de Vrind-de Jong and de Vrind 1997; Young et al. 1999; Marsh 2000; González 2000; Paasche 2002; Baeuerlein 2003), and the molecules responsible for the acquisition and intracellular transport of Ca2+, HCO3 − and CO3 2−, and in the precipitation of CaCO3 remain to be identified.
However, a whole genome assembly for E. huxleyi (strain CCMP1516) has been publicly released by the Joint Genome Institute (available at www.doe.jgi.gov), a growing number of expressed sequence tags (EST) resources for this species are now available (Wahlund et al. 2004; Quinn et al. 2006; von Dassow et al. 2009), and candidate genes likely to be important for biomineralization can now be identified by homology to known eukaryotic proteins involved in the processing of Ca2+ and CO2/HCO3 −/CO3 2−.
In the present study, we chose E. huxleyi (Lohmann) Hay and Mohler, the most abundant calcifying phytoplankton on Earth (Westbroek et al. 1993) to investigate the effect of atmospheric CO2 emission scenarios expected by the end of this century (IPCC 2007) on calcification process and underlying cellular mechanisms. We assessed the growth and calcification rate of a calcifying strain of this species in response to pCO2/pH variations. In parallel, molecular targets were followed for their gene expression using quantitative PCR.
We focused on two classes of proteins tightly involved in cellular pH and/or carbonate chemistry regulation (e.g. carbonic anhydrase and Cl−/HCO3 − anion exchanger family). We studied two classes of carbonic anhydrase (CA) out of five known (α, β, γ, δ, and ζ) and their role in E. huxleyi cells subjected to lower pH. Carbonic anhydrases are ubiquitous metalloenzymes that catalyze the reversible hydration of carbon dioxide into bicarbonate and play different roles in physiological processes such as photosynthesis, respiration, pH homeostasis and ion transport.
We also investigated the homologs of Cl−/bicarbonate exchanger solute carrier family 4 proteins (SLC4), well known for their roles in intracellular pH regulation in animal cell (Romero et al. 2004) and recently described as highly specific to calcifying cells of E. huxleyi (von Dassow et al. 2009). According to von Dassow et al. (2009) study, one of the SLC4 Cl−/bicarbonate transcript (cluster GS05051) was represented by 7/0 reads for calcifying (2 N) cells compared to non-calcifying (N) cells.
Based on the decrease in calcification (e.g. decrease in PIC) observed in some coccolithophore cultures subjected to pCO2 increase (Riebesell et al. 2000; Zondervan et al. 2001; Sciandra et al. 2003; Langer et al. 2006, 2009; Feng et al. 2008; Müller et al. 2010; Ridgwell et al. 2009), representative genes of two more protein classes were then investigated. The protein GPA was chosen since it was previously found associated with coccolith polysaccharides and displays Ca2+-binding activity (Corstjens et al. 1998).
We then chose to specifically examine a Ca2+-transporter-related gene. Ca2+ ion is not only a regulatory agent in physiological processes but also the primary cation used in biomineralized structures. While Ca2+ transporters and specifically the voltage-gated ion channel proteins are described in detail for vertebrates (Dolphin 2009), little is known about such transporters in the protist E. huxleyi. However, as in all biomineralization processes, either intracellular or extracellular, the primary event is the entry of Ca2+ ions at the cell membrane level. Thus, we hypothesized that those genes might be involved in calcification of E. huxleyi as it has been previously shown in the scleractinian coral Stylophora pistillata (Zoccola et al. 1999) and in calcification process in general.
In the present study, the hypothetical roles of the genes of interest in calcification in relation to the expression response to pH/pCO2 variations and perspectives for the future of coccolithophores in a high CO2 world are discussed.
Materials and methods
Culture condition and sampling
Diploid (2 N) cells of Emiliania huxleyi strain RCC1216 (Tasman sea; 42°18′S–169°50′W) were provided by the Algobank culture collection, Caen, France (http://www.sb-roscoff.fr/Phyto/RCC). Many E. huxleyi strains lose the capacity to calcify in culture, and cultures often contain a mix of non-calcified and calcified cells complicating interpretations. Haploid and diploid life stages of the studied strain (RCC1217/RCC1216) were first characterized on a flow cytometer. Two distinct groups were identified in cytograms according to their nucleic acid fluorescence and side scatter. The composition of the experimental culture was then confirmed to be mainly diploid. RCC1216 was chosen because a wealth of ESTs is available from this strain and it exhibits high calcification under standard culture conditions. Cultures were maintained in K/2 (-Si, -Tris) medium prepared from filter-sterilized seawater (Keller et al. 1987) at 17°C under a 14 h light: 10 h dark photoperiod with cool white fluorescent light at 150 μmol photons m−2 s−1, with a salinity of 38 0/00.
Experimental setup
Two 10-l glass bottles (control and experimental treatments) were filled with sterile culture medium and maintained at 17°C using a thermostated water bath. They were bubbled for 2 h with ambient air (control, ambient pCO2) or a mixture CO2-free air (generated by the use of soda lime) and pure CO2 stabilized at the desired partial pressure of 760 ppm (experimental treatment, high pCO2) by a mass flow controller (GFC, Aalborg) coupled with an infrared gas analyzer (LICOR Li-6252), respectively. pH, salinity and total alkalinity (TA) were measured to check the pCO2 in both treatments. The final pCO2 values were 440 and 770 ppm in the control and the experimental treatments, respectively. Once the desired pCO2 was reached, triplicate 2-l Nalgene bottles were filled up with each medium without headspace. An inoculum of 50 cells ml−1 (calculated from the stock culture) was added, and the 6 bottles were sealed with Teflon tape to avoid gas exchange between the medium and the atmosphere (Langer et al. 2006). Replicates were transferred to an incubation chamber and kept under the conditions described earlier (see Culture condition and sampling section) during all the experimental period. The cells were harvested at around 50,000 ± 10,000 cells ml−1 in order to work with low cell densities ensuring well-controlled experimental conditions and the biomass necessary for a reliable analysis. The sampling was performed after an 8-day incubation period at 0900 h (90 min after the beginning of the light period) for all 6 bottles.
Cell density and growth rate
Cell density was checked daily (10.00 a.m.) from day 3, using 500 μl of sample on a flow cytometer (FACSCalibur, BD Biosciences). Coccolithophores were detected by their red autofluorescence in the FL3 channel.
For determination of the growth rate (μ), samples for cell density were taken at the beginning and at the end of experiment. Growth rate (μ) was calculated as: μ = (lnC1−lnC0)Δt−1 where C0 and C1 are the cell concentrations at the beginning (inoculation time) and at the end of experiment (harvesting time), respectively, and Δt is the duration of incubation in days.
Carbonate chemistry measurements
The carbonate system of the experiment was monitored by measuring total alkalinity (TA), pHT, temperature and salinity in the cultures. Triplicate 25 ml samples were collected for total alkalinity at the beginning, prior to inoculation, and at the end of the experiments (harvesting time). They were immediately filtered onto 0.2-μm filters and analyzed potentiometrically by a custom-made titrator built with a Metrohm pH electrode and a 665 Dosimat titrator. TA was calculated using a Gran function applied to the pH values ranging from 3.5 to 3.0 as described by Dickson et al. (2007). Titrations of an alkalinity standard, provided by A. G. Dickson (batch 80), were within 0.7 μmol kg−1 of the nominal value (SD = 2.6 μmol kg−1; N = 8). According to Brewer and Goldman (1976), 1 μM EDTA added to a phytoplankton culture to maintain Fe in solution contributes about 2 μeq to the alkalinity. In our case, the 125 nM EDTA should contribute about 0.2 μeq to the alkalinity in the medium and can thus be considered as negligible.
pHT was measured on 20 ml samples using a pH meter (Metrohm, 826 pH mobile) with a glass electrode (Ecotrode, 6.0262.100 Metrohm) calibrated on the total scale using Tris/HCl and 2-aminopyridine/HCl buffer solutions with a salinity of 38 at a temperature of 17°C. pCO2, Ωcalcite and other parameters of the carbonate system were calculated from given TA and pH using the R package seacarb (Lavigne et al. 2008). The carbonate system, at the beginning and at the end of the incubation period (8 days), is described in Table 1.
Particulate inorganic (PIC) and organic (POC) carbon measurements
Triplicate samples (~150 μg C per filter) were filtered onto pre-combusted (4 h, 400°C) glass fiber filters (Whatman GF/F), dried at 60°C overnight and stored in a desiccator pending analysis. For POC measurements, the inorganic carbon was removed from the filters before the analysis by adding 25% HCl (Nieuwenhuize et al. 1994). Cell content for total particulate carbon (TPC) and for particulate organic carbon (POC) (pg cell−1) was subsequently measured on a Thermo Electron Flash EA 1112 Analyzer as described by Nieuwenhuize et al. (1994). Particulate inorganic carbon (PIC) (pg cell−1) was calculated as the difference between TPC and POC. Particulate inorganic carbon production, i.e. calcification rate (PPIC, pg PIC cell−1 d−1) was calculated according to: PPIC = μ × (cellular inorganic carbon content in pg PIC per cell). Particulate organic carbon production (PPOC, pg POC cell−1 d−1) was calculated according to: PPOC = μ × (cellular organic carbon content in pg POC per cell) (Riebesell et al. 2000).
Quantitative reverse transcriptase-polymerase chain reaction (q-RT–PCR)
RNA extraction—Total RNA was isolated from coccolithophores with Trizol reagent (Invitrogen, La Jolla, CA) according to the suggested protocol. Five hundred milliliter of medium from each bottle was collected by gentle filtration on polycarbonate filter of 1 μm (Whatman) and resuspended in 1 ml of Trizol. Two successive chloroform (≥99%) steps in 200 μl were carried out to precipitate proteins and DNA. RNA was finally precipitated in 500 μl isopropanol (≥99%). The pellets were washed in 75% ethanol and resuspended in RNase-free water. The RNA quality was checked on 1% agarose (w:v) non-denaturing gels and the purity determined using a Nanodrop spectrophotometer (Nanodrop 3300, Thermo scientific). All samples presented ribosomal RNA bands with no sign of degradation. RNA samples were treated with DNase (1U μl−1, Fermentas) and quantified using a RiboGreen RNA Quantification Kit (Molecular Probes). Total RNA concentration was adjusted to a final concentration of 100 ng μl−1 in all samples, and the reverse transcription was carried out using the Affinity Script qPCR cDNA kit (Stratagene). Negative controls (same reagents mix without reverse transcriptase) were prepared simultaneously and run on each plate for each primer pairs to ascertain that no DNA contamination occurred (Ct values were >40 cycles). No template controls were also run in parallel on each plate.
Transcript levels were derived from the accumulation of SYBR green fluorescence measured with a Light Cycler 480 (Roche). The PCR conditions were as follows: 1× SYBR green mix (Roche, Cat. nb: 04707516001), 500 nM primers and 1 μl (100 ng) of cDNA in a total volume of 20 μl. Each sample was run in triplicate (mean ± SD < 0.2). The dissociation curves showed a single amplification product and no primer dimer. For each primer pairs, the amplification efficiency (E) was determined on a 5 points 10-time dilution series of 100 ng cDNA extracted from the two tested conditions (control and experimental pCO2) to check for primer specificity. The reaction efficiencies had values between 80 and 100% with a corresponding amplification factor between 1.8 and 2.0, respectively, for all primer combinations. This value allows for a transformation of the observed changes in cycle threshold (CT).
RNA transcription levels were determined by the method of direct comparison of CT values between target genes and a reference gene. Several genes from E. huxleyi strain CCMP1516 (JGI, USA) commonly used as housekeeping genes (HKG) (e.g. actin (JGI, ID 226687), β-tubulin (JGI, ID 451245) and RPLP0 (JGI, ID 456254)) were tested for their expression stability in experimental samples using the program geNorm (Vandesompele et al. 2002). While none of them was stable enough to normalize the data, calmodulin (JGI, ID 442625) was identified as the most stable gene and used further to normalize the data by the ΔΔCt method (Livak and Schmittgen 2001). Data were then transformed into linear form by: 2−DDCT where −DDCT = (CtTarget−CtHKG)Tx−(CtTarget−CtHKG)T0. Data were analyzed using one-way analyses of variance (ANOVA). Since all the steps from RNA extraction to RT qPCR efficiency have been checked for accuracy, high standards deviations (SD) reported in Fig. 3 were mainly attributed to biological variability in experimental batch cultures.
Genes of interest and primer design
The sequences of 4 of the genes investigated here (α- and γ_CA, Ca2+-channel and gpa) were obtained from E. huxleyi strain CCMP1516 genome portal (http://shake.jgi-psf.org/Emihu1/Emihu1.home.html). The transcripts that encode Cl−/HCO3 − exchanger homologs (SLC4 family) were annotated from the Sanger reads of E. huxleyi (strains RCC1216/RCC1217) cDNA libraries (von Dassow et al. 2009). Up to 7 homologs have been investigated (GS00443, GS02476, GS12371, GS03121, GS05051, GS09941, GS05509) but only 6 are presented in this study (GS00443 was weakly represented and not significantly detected by qPCR).
In order to characterize the coding sequences (partial or complete) chosen as part of this study, the amino acid (aa) sequences (α- and γ_CA, Ca2+-channel and GPA) or nucleotide sequences (Cl−/HCO3 − exchanger homologs) were blasted to UniProt/Swiss-Prot databases (Consortium U 2009) and NCBI/CDD (Conserved Domains database) (Marchler-Bauer et al. 2009). The characteristics of the given sequences are detailed in Tables 2, 3.
qPCR primer sequences were designed using the Primer3 software to have a G + C content ranging from 50 to 60% and C’s > G’s 3 identical dNTPs in a row at the 3′ ends to avoid self complementarities of the primer sequence. Primers were chosen to generate equivalent amplicon lengths (see Table 4). The melting temperature of the primers was set at 58°C. The qPCR products were sequenced (MWG, Germany) and all matched the anticipated product. For PCR products obtained with primers designed from E. huxleyi strain CCMP1516 (e.g. α- and γ_CA, Ca2+-channel and GPA), sequences from both strains (CCMP1516 and RCC1216) were aligned and showed 100% identity.
Results and discussion
While previous molecular studies on E. huxleyi dealt with identification of genes that are associated with the calcification mechanism (Quinn et al. 2006; Wahlund et al. 2004; Nguyen et al. 2005; von Dassow et al. 2009), our experiment is the first to investigate gene expression in response to CO2-driven ocean acidification. Our approach provides new elements on the molecular and physiological role of genes of interest in calcification and helps understand the diverse response of coccolithophores to projected ocean acidification.
Physiological and biochemical response to decreasing pH
The experimental setup was designed following recommendations of best practices (Riebesell et al. 2010), and batch cultures were used as many other previous studies (Riebesell et al. 2000; Zondervan et al. 2001, 2002; Langer et al. 2006, 2009; Iglesias-Rodriguez et al. 2008), in order to ensure that data comparison between studies is possible. The manipulation of the carbonate system was achieved by bubbling the culture medium with CO2 and/or air before the inoculation, and the experiment was consequently performed in a closed system avoiding gas exchanges with the atmosphere. As in the natural environment, this method involves changes in pCO2, DIC and pH, while TA remains constant (Gattuso and Lavigne 2009). The stress caused to the cultures by the air bubbling and consequent variability of the response to tested parameters are eliminated, and the shift in carbonate parameters due to cell activity is negligible. Consequently, any change during the experiment can exclusively be attributed to physiological changes in response to the CO2 perturbation (Fiorini 2010).
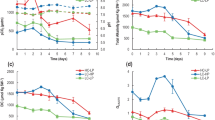
In the past few years, parameters such as growth rate and organic and inorganic carbon production have been widely investigated in calcifiers in order to predict the impact of ocean acidification (Buitenhuis et al. 1999; Clark and Flynn, 2000; Riebesell et al. 2000; Rost et al. 2002; Sciandra et al. 2003; Iglesias-Rodriguez et al. 2008: Shi et al. 2009; Barcelos e Ramos et al. 2010; Müller et al. 2010; Langer et al. 2009). In this study, the response regarding those parameters is in agreement with the diverse responses already described for E. huxleyi strains in the literature. We found a minor effect of elevated pCO2 on the physiology of E. huxleyi RCC1216. Cell density was not significantly changed at elevated pCO2 (Student t test P < 0.1) (Fig. 1), and growth rate remained unchanged with μ = 0.79 ± 0.02 and 0.76 ± 0.02 for cultures subjected to control and elevated pCO2, respectively (Student t test P < 0.1).
Likewise, no significant change in production of particulate organic (PPOC) and inorganic (PPIC) carbon (Fig. 2a) (Student t test P < 0.1) and PIC/POC ratio (Fig. 2b) (Student t test P < 0.4) was observed in cultures subjected to low or high pCO2. A recent work by Langer et al. (2009), dealing with the response of E. huxleyi strain RCC1216 to changing seawater carbonate chemistry, showed both a decrease in PIC cellular content and production in cultures subjected to a pCO2 of 729 μatm. The reasons for the discrepancy might relate to differences in culture conditions. Whereas cultures were pre-adapted to experimental conditions for 12 generations by Langer et al. (2009), we only subjected our cultures to an 8-day treatment without acclimation period.
Molecular responses to decreasing pH
In this study, we have used gene expression profiling to explore some molecular mechanisms that may underlie tolerance to future ocean acidification conditions. The genes investigated were related to the Ca2+ metabolism or speciation and DIC transport (see Table 2). The expression of up to 11 genes was followed and mainly showed no significant response to elevated pCO2/lower pH (Fig. 3).
Although all of the genes of interest were previously described as potentially associated with the biomineralization process in a wide range of organisms, from pelagic coccolithophores (Wahlund et al. 2004; Dyhrman et al. 2006; Soto et al. 2006; Quinn et al. 2006; Richier et al. 2009) to benthic invertebrates (Zoccola et al. 1999; Moya et al. 2008), this is the first time that genes related to the SLC4 family have been investigated for their role in the carbonate chemistry of marine calcifiers and their response to environmental threats (e.g. ocean acidification). The family of SLC4 anion exchanger (AE) proteins includes the Na+-independent Cl−/HCO3 − exchanger that is critical for the regulation of several physiological processes including intracellular pH (pHi) and the HCO3 −/CO3 2− balance in eukaryotic cells (Alper et al. 2001; Alper 2009; Romero et al. 2004). In E. huxleyi, an inhibitor-based study has indicated the involvement of the Cl−/HCO3 − exchangers in DIC uptake (Herfort et al. 2002), and von Dassow et al. (2009) recently suggested that SLC4 homolog might function to maintain optimal balance of pH and carbonate/bicarbonate in the coccolith deposition vesicle for calcification. However, SLC4-like homolog ability to transport either HCO3 − or CO3 2− in E. huxleyi is still unknown (Mackinder et al. 2010). In the present study, no significant variation in the Cl−/HCO3 − exchanger homolog’s gene expression was observed under tested conditions (Fig. 3; ANOVA one-way, P < 0.5). Those results suggest either undetectable or no effect of the tested pH/pCO2 perturbation on targeted genes. In fact, the low variations in the carbonate system (i.e. ΔHCO3 −/CO3 2−) highlighted in our experiment (see Table 1) could explain the unchanged Cl−/HCO3 − exchanger gene expression. In sea urchin larvae subjected to similar pCO2 condition, unchanged mRNA transcript levels of the Cl−/HCO3 − exchanger were also reported (Todgham and Hofmann 2009).
Looking further into genes related to DIC transport proteins, the expression of α- and γ-CA genes was investigated as part of this study. Information on the molecular characterization of CA is scarce in phytoplankton, and especially in coccolithophores. The involvement of these two CAs in biomineralization has yet to be discussed but given the role of CA in acid/base compensation, it is probable that one or more of them may be regulated by the acid–base imbalance that could have resulted from the decrease in pH. Despite up to 12 CA transcripts recently identified in E. huxleyi by von Dassow et al. (2009), little information is available about the localization and role of these genes in coccolithophores. A first attempt to characterize CA isoforms in E. huxleyi was performed by Soto et al. (2006) who speculated on a location for γ-EhCA2 protein in the coccolith vesicle with a 25-fold up-regulation in γ-EhCA2 transcripts under calcifying versus non-calcifying condition. The role of γ-CA isoform in calcification was also supported by previous studies with up-regulated transcripts in cultures where calcification was stimulated by phosphate-depletion (Quinn et al. 2006) and significantly higher during the light period in calcifying cells (RCC1216 strain) compared to non-calcifying ones (RCC1217) (Richier et al. 2009).
In the present study, the CA sequences were searched against databases for their conserved domains (see Table 2). A conserved domain homolog to an “alpha_CA_procaryotic like” carbonic anhydrase was detected in α-CA. In this sub-family, the enzyme has been reported to be part of the organic matrix layer in shells. Other members of this family may be involved in maintaining pH balance, in facilitating transport of CO2 or H2CO3, or in sensing carbon dioxide levels in the environment. We thus deliberately chose here to analyze γ-CA isoform, for the reasons outlined earlier, and α-CA isoform for its widespread distribution in several kingdoms of life (vertebrates, invertebrates, bacteria, and some chlorophytes) and its role in biomineralization of benthic organisms (Moya et al. 2008). We showed that α- and γ-CA genes were down-regulated when exposed to decreasing pH resulting in a fold change of 2.3 and 3.8, respectively (ANOVA one-way, P < 0.05) (Fig. 3). A previous study on E. huxleyi intracellular CA activity showed no clear trend with increasing pCO2 (from 36 ppmv up to 1,800 ppmv) (Rost et al. 2003). However, the measurements in that study did not discriminate between CA isoform classes and it might be that the regulation of CA genes is class specific.
Additionally, it has been previously suggested that the CA enzymes and SLC4 anion exchangers may interact (Vince and Reithmeier, 2000; Sterling et al. 2001, 2002; Morgan et al. 2007). In mammalian cell lines, the cytoplasmic carboxy terminal of AE1 has a carbonic anhydrase II (CAII) binding site that upon inhibition reduces AE1-mediated Cl−/HCO3 − exchange by 50–60% (Sterling et al. 2001). Carbonic anhydrase IV (CAIV) interaction sites have also been identified on the extracellular surface of AE1 isoform. According to the authors, CAII and IV would increase HCO3 − transport by altering localized HCO3 − levels enhancing the HCO3 − concentration gradient (McMurtrie et al. 2003). A similar function may occur in coccolithophores with CA interacting with the Cl−/HCO3 − exchanger facilitating the conversion of HCO3 − into CO2 at the cytosolic face of the plasma membrane decreasing the local concentration of HCO3 − at the cytosolic transport site (Mackinder et al. 2010). In our study, we could speculate that increasing pCO2 inhibits both α- and γ-CA genes transcription and consequently the activity of their relative proteins. Thus, no interactions with SLC4 homologs would occur, which is reflected by unchanged Cl−/HCO3 − exchanger transcript level under experimental condition. In the same way, the unchanged Ca2+-channel (CAC) and gpa transcript level, in response to tested conditions, would suggest no reduced capacity of the protein to transport or bind Ca2+ to the sites of calcification and supports the unchanged calcification rate observed in the tested cultures. However, the regulation of gene of interest related proteins was not investigated as part of this study. Simultaneous analyses of both transcripts and corresponding proteins are required to conclude on any proteins regulation and function.
In conclusion, all the results shown by our study constitute new elements in molecular exploration of genes involved in E. huxleyi early response to an acidifying world. No major physiological changes were observed in the chosen strain in response to ocean acidification and only CA isoforms, among the tested genes, appeared significantly regulated under the experimental condition. However, no significant variation in expression of most of the genes might either suggest (1) no major effect of the near future pCO2 condition in the ocean on the tested strain or (2) no direct role of the targeted genes in early response to high pCO2/low pH. An exhaustive investigation into E. huxleyi transcriptome would be required to identify all the genes/cellular mechanisms involved in response to pCO2/pH variation.
Nonetheless, the fact high pCO2-treatment did not induce major molecular and physiological changes in this calcified phytoplankton suggests that it may have the capacity to adapt to future ocean acidification.
References
Alper SL (2009) Molecular physiology and genetics of Na+-Independent SLC4 anion exchangers. J Exp Biol 212:1672–1683
Alper SL, Chernova MN, Stewart AK (2001) Regulation of Na+-Independent Cl−/HCO3 − exchangers by pH. JOP J Pancreas 2:171–175
Baeuerlein E (2003) Biomineralization of unicellular organisms: an unusual membrane biochemistry for the production of inorganic nano- and microstructures. Angew Chem Int Ed 42:614–641
Barcelos e Ramos J, Muller MN, Riebesell U (2010) Short-term response of the coccolithophore Emiliania huxleyi to abrupt changes in seawater carbon dioxide concentrations. Biogeosciences 7:177–186
Brewer PG, Goldman JC (1976) Alkalinity changes generated by phytoplankton growth. Limnol Oceanogr 21:108
Buitenhuis ET, De Baar HJW, Veldhuis MJW (1999) Photosynthesis and calcification by Emiliania huxleyi (Prymnesiophyceae) as a function of inorganic carbon species. J Phycol 35:949–959
Clark DR, Flynn KJ (2000) The relationship between the dissolved inorganic carbon concentration and growth rate in marine phytoplankton. Proc R Soc Lond 267:953–959
Consortium U (2009) The Universal Protein Resource (UniProt). Nucleic Acids Res 37:D169–D174
Corstjens PLAM, van der Kooij A, Linschooten C, Brouwers GJ, Westbroek P, de Vrind-de Jong EW (1998) GPA, a calcium-binding protein in the coccolithophorid Emiliania huxleyi (Prymnesiophyceae). J Phycol 34:622–630
de Vrind-de Jong EW, de Vrind JPM (1997) Algal deposition of carbonates and silicates. In: Banfield JF, Nealson KH (eds) Geomicrobiology: interactions between microbes and minerals. Mineralogical Society of America, Washington, DC, pp 267–307
Dickson AG, Sabine CL, Christian JR (eds) (2007) Guide to best practices for ocean CO2 measurements, PICES Special Publication 3, p 191
Dolphin AC (2009) Calcium channel diversity: multiple roles of calcium channel subunits. Curr Opin Neurobiol 19:237–244
Dyhrman ST, Haley ST, Birkeland SR, Wurch LL, Cipriano MJ, McArthur AG (2006) Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi. Appl Environ Microbiol 72:252–260
Engel A, Szlosek J, Abramson L, Liu Z, Lee C (2009) Investigating the effect of ballasting by CaCO3 in Emiliania huxlei: I. Formation, settling velocities and physical properties of aggregates. Deep Sea Res II 56:1396–1407
Feng Y, Warner ME, Zhang Y, Sun J, Fu FX, Rose JM, Hutchins DA (2008) Interactive effects of increased pCO2, temperature and irradiance on the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae). Eur J Phycol 43:87–98
Fiorini S (2010) Effect of elevated CO2 partial pressure and temperature on calcifying phytoplankton (coccolithophores). Ph.D. dissertation, Pierre and Marie Curie University, Paris, France
Gattuso J-P, Lavigne H (2009) Technical note: approaches and software tools to investigate the impact of ocean acidification. Biogeosciences 6:2121–2133
González EL (2000) The calcifying vesicle membrane of the coccolithophore. In: Baeuerlein E (ed) Biomineralization: from biology to biotechnology and medical application. Wiley-VCH, Weinheim, pp 269–283
Halpern BS, Walbridge S, Selkoe KA, Kappel CV, Micheli F, D’Agrosa C et al (2008) A global map of human impact on marine ecosystems. Science 319:948–952
Herfort L, Thake B, Roberts J (2002) Acquisition and use of bicarbonate by Emiliania huxleyi. New Phytol 156:427–436
Iglesias-Rodriguez MD, Halloran PR, Rickaby RE, Hall IR, Colmenero-Hidalgo E, Gittins JR et al (2008) Phytoplankton calcification in a high-CO2 world. Science 320:336–340
Keller MD, Selvin RC, Claus W, Guillard RRL (1987) Media for the culture of oceanic ultraphytoplankton. J Phycol 23:633–638
Langer G, Geisen M, Baumann KH, Klas J, Riebesell U, Thoms S, Young JR (2006) Species-specific responses of calcifying algae to changing seawater carbonate chemistry. Geochem Geophys Geosyst 7:Q09006
Langer G, Nehrke G, Probert I, Ly J, Ziveri P (2009) Strain-specific responses of Emiliania huxleyi to changing seawater carbonate chemistry. Biogeosciences 6:2637–2646
Lavigne H, Proye A, Gattuso J-P (2008) seacarb 2.0, an R package to calculate parameters of the seawater carbonate system. Available at http://cran.rproject.org/web/packages/seacarb/index.html
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-DDCT method. Methods 25:402–408
Mackinder L, Wheeler G, Schroeder D, Riebesell U, Brownlee C (2010) Molecular mechanisms underlying calcification in coccolithophores. Geomicrobiology 27:585–595
Marchler-Bauer A et al (2009) CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res 37:205–210
Marsh ME (2000) Polyanions in the CaCO3 mineralization of coccolithophores. In: Baeuerlein E (ed) Biomineralization: from biology to biotechnology and medical application. Wiley-VCH, Weinheim, pp 251–268
McMurtrie HL, Cleary HJ, Alvarez BV, Loiselle FB, Sterling D, Morgan PE, Johnson DE, Casey JR (2003) The bicarbonate transport metabolon. In: 6th international conference on carbonic anhydrases, Taylor & Francis Ltd, Bratislava, Slovakia, pp 231–236
Milliman JD (1993) Production and accumulation of calcium carbonate in the ocean—budget of a nonsteady state. Global Biogeochem Cycles 7:927–957
Morgan PE, Pastoreková S, Stuart-Tilley AK, Alper SL, Casey JR (2007) Interactions of transmembrane carbonic anhydrase, CAIX, with bicarbonate transporters. Am J Physiol Cell Physiol 293:738–748
Moya A, Tambutté S, Bertucci A, Tambutté E, Lotto S, Vullo D et al (2008) Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J Biol Chem 283:25475–25484
Müller MN, Schulz KG, Riebesell U (2010) Effects of long-term high CO2 exposure on two species of coccolithophores. Biogeosciences 7:1109–1116
Nguyen B, Bowers RM, Wahlund TM, Read BA (2005) Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Appl Environ Microbiol 71:2564–2575
Nieuwenhuize J, Maas YEM, Middelburg JJ (1994) Rapid analysis of organic carbon and nitrogen in particulate materials. Mar Chem 44:217–224
Orr JC, Fabry VJ, Aumont O, Bopp L, Doney SC, Feely RA et al (2005) Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437:681–686
Paasche E (2002) A review of the coccolithophore Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification–photosynthesis interactions. Phycologia 40:503–529
Quinn P, Bowers RM, Zhang X, Wahlund TM, Fanelli MA, Olszova D, Read BA (2006) cDNA microarrays as a tool for identification of biomineralization proteins in the coccolithophore Emiliania huxleyi (Haptophyta). Appl Environ Microbiol 72:5512–5526
Richier S, Kerros ME, de Vargas C, Hamaraty L, Falkowski PG, Gattuso J-P (2009) Light-dependent transcriptional regulation of genes of biogeochemical interest in the diploid and haploid life cycle stages of Emiliania huxleyi. Appl Environ Microbiol 75:3366–3369
Ridgwell A, Schmidt DN, Turley C, Brownlee C, Maldonado MT, Tortell P, Young JR (2009) From laboratory manipulations to earth system models: scaling calcification impacts of ocean acidification. Biogeosciences 6:2611–2623
Riebesell U, Zondervan I, Rost B, Tortell PD, Morel FFM (2000) Reduced calcification of marine plankton in response to increased atmospheric CO2. Nature 407:364–367
Riebesell U, Fabry VJ, Hansson L, Gattuso J-P (eds) (2010) Guide to best practices for ocean acidification research and data reporting. Publications Office of the European Union, Luxembourg, p 260
Romero MF, Fulton CM, Boron WF (2004) The SLC4 family of HCO3 − transporters. Eur J Physiol 447:495–509
Rost B, Zondervan I, Riebesell U (2002) Light-dependent carbon isotope fractionation in the coccolithophore Emiliania huxleyi. Limnol Oceanogr 47:120–128
Rost B, Riebesell U, Burkhardt S (2003) Carbon acquisition of blooming marine phytoplankton. Limnol Oceanogr 48:55–67
Sabine CL, Feely RA, Gruber N, Key RM, Lee K, Bullister JL et al (2004) The oceanic sink for anthropogenic CO2. Science 305:367–371
Sciandra A, Harlay J, Lefevre D, Lemee R, Rimmelin P, Denis M, Gattuso J-P (2003) Response of coccolithophorid Emiliania huxleyi to elevated partial pressure of CO2 under nitrogen limitation. Mar Ecol Prog Ser 161:111–122
Shi D, Xu Y, Morel FMM (2009) Effects of the pH/pCO2 control method on medium chemistry and phytoplankton growth. Biogeosciences 6:1199–1207
Soto AR, Zheng H, Shoemaker D, Rodriguez J, Read BA, Wahlund TM (2006) Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi. Appl Environ Microbiol 72:5500–5511
Sterling D, Reithmeier RA, Casey JR (2001) A transport metabolon: functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J Biol Chem 276:47886–47894
Sterling D, Alvarez BV, Casey JR (2002) The extracellular component of a transport metabolon: extracellular loop 4 of the human AE1 Cl−/HCO3 − exchanger binds carbonic anhydrase IV. J Biol Chem 277:25239–25246
Todgham AE, Hofmann GE (2009) Transcriptomic response of sea urchin larvae Strogylocentrotus purpuratus to CO2-driven seawater acidification. J Exp Biol 212:2579–2594
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:34.1–34.11
von Dassow P, Ogata H, Probert I, Wincker P, Da Silva C, Audic, S, Claverie J-M, de Vargas C (2009) Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol. doi:10.1186/gb-2009-10-10-r114
Vince JW, Reithmeier RA (2000) Identification of the carbonic anhydrase II binding site in the Cl_/HCO _3 anion exchanger AE1. Biochemistry 39:5527–5533
Wahlund TM, Hadaegh AR, Clark R, Nguyen B, Fanelli M, Read BA (2004) Analysis of expressed sequence tags from calcifying cells of marine coccolithophorid (Emiliania huxleyi). Mar. Biotechnol (NY) 6:278–290
Westbroek P, Brown CW, Vanbleijswijk J, Brownlee C, Brummer GJ, Conte M, Egge J, Fernandez E, Jordan R, Knappertsbusch M (1993) A model system approach to biological climate forcing - the example of Emiliania huxleyi. Glob Planet Change 8:27–46
Young JR, Davis SA, Bown PR, Mann S (1999) Coccolith structure and biomineralization. J Struct Biol 126:195–215
Zoccola D, Tambutté E, Senegas-Balas F, Michiels JF, Failla JP, Jaubert J, Allemand D (1999) Cloning of a calcium channel a1 subunit from the reef-building coral, Stylophora pistillata. Gene 227:157–167
Zondervan I, Zeebe RE, Rost B, Riebesell U (2001) Decreasing marine biogenic calcification: a negative feedback on rising atmospheric pCO2. Glob. Biogeochem Cycles 15:507–551
Zondervan I, Rost B, Riebesell U (2002) Effect of CO2 concentration on the PIC/POC ratio in the coccolithophore Emiliania huxleyi grown under light-limiting conditions and different day lengths. J Exp Mar Biol Ecol 272:55–70
Acknowledgments
We thank Cornelia Maier and JinWen Liu for providing access to mass flow controllers and assistance to set up the high pCO2 experiment. We also thank Steeve Comeau for technical support with measurements of pH and total alkalinity. We are also grateful to Anna Macey for her help with the English. This is a contribution to the “European Project on Ocean Acidification” (EPOCA) which receives funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 211384. We are also grateful to several anonymous reviewers that significantly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by U. Sommer.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Richier, S., Fiorini, S., Kerros, ME. et al. Response of the calcifying coccolithophore Emiliania huxleyi to low pH/high pCO2: from physiology to molecular level. Mar Biol 158, 551–560 (2011). https://doi.org/10.1007/s00227-010-1580-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1580-8