Abstract
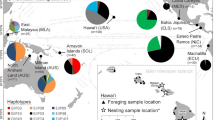
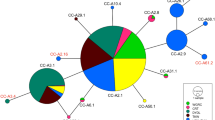
The green turtle (Chelonia mydas) nesting population at Tortuguero, Costa Rica, is the largest nesting aggregation in the Atlantic, by at least an order of magnitude. Previous mitochondrial DNA (mtDNA) surveys based on limited sampling (n = 41) indicated low genetic diversity and low gene flow with other Caribbean nesting colonies. Furthermore, a survey of nuclear DNA diversity invoked the possibility of substructure within the Tortuguero rookery. To evaluate these characteristics, mtDNA control region sequences were determined for green turtles nesting at Tortuguero in 2001 (n = 157) and 2002 (n = 235). The increased sample revealed three additional haplotypes; five haplotypes are now known for Tortuguero female green turtles. Analyses of molecular variance indicated that there was no significant spatial population structure along the 30-km nesting beach. In addition, no temporal population structure was detected either between the two nesting seasons or within the nesting season. As a result of the larger sample size and additional haplotypes, estimates of genetic separation among Caribbean nesting colonies have changed and the concordance of phylogenetic and phylogeographic patterns reported in the past for green turtles in the Greater Caribbean has weakened. The five haplotypes from Tortuguero represent 36% of the haplotypes identified in green turtle nesting aggregations in the Greater Caribbean and 17% of the haplotypes known to occur in nesting or foraging aggregations in the Greater Caribbean. Haplotype diversity (0.16) and nucleotide diversity (0.0034) for the Tortuguero population are substantially lower than those for the combined rookeries in the Greater Caribbean (0.44 and 0.0078, respectively). Although comprehensive evaluation of regional genetic diversity requires nuclear DNA data, our study indicates that conserving genetic diversity in Caribbean green turtles will require careful management of the smaller rookeries in addition to the Tortuguero rookery.


Similar content being viewed by others
References
Bass AL, Witzell WN (2000) Demographic composition of immature green turtles (Chelonia mydas) from the east central Florida coast: evidence from mtDNA markers. Herpetologica 56:357–367
Bass AL, Lagueux CJ, Bowen BW (1998) Origin of green turtles, Chelonia mydas, at “sleeping rocks” off the northeast coast of Nicaragua. Copeia 1998:1064–1069
Bjorndal KA, Wetherall JA, Bolten AB, Mortimer JA (1999) Twenty-six years of green turtle nesting at Tortuguero, Costa Rica: an encouraging trend. Conserv Biol 13:126–134
Bolker B, Okuyama T, Bjorndal K, Bolten A (2003) Sea turtle stock estimation using genetic markers: accounting for sampling error of rare genotypes. Ecol Appl 13:763–775
Bolten AB (2003) Variation in sea turtle life history patterns: neritic vs. oceanic developmental stages. In: Lutz PL, Musick J, Wyneken J (eds) The biology of sea turtles, vol II. CRC Press, Boca Raton, FL, pp 243–257
Bolten AB, Bjorndal KA, Martins HR, Dellinger T, Biscoito MJ, Encalada SE, Bowen BW (1998) Transatlantic developmental migrations of loggerhead sea turtles demonstrated by mtDNA sequence analysis. Ecol Appl 8:1–7
Bowen BW (1995) Molecular genetic studies of marine turtles. In Bjorndal KA (ed) Biology and conservation of sea turtles (revised edition). Smithsonian Institution Press, Washington, DC, pp 585–587
Bowen BW (2003) What is a loggerhead turtle? The genetic perspective. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution Press, Washington, DC, pp 7–27
Bowen BW, Karl SA (1997) Population genetics, phylogeography, and molecular evolution. In: Lutz PL, Musick JA (eds) The biology of sea turtles. CRC Press, Boca Raton, FL, pp 29–50
Bowen BW, Meylan AB, Ross JP, Limpus CJ, Balazs GH, Avise JC (1992) Global population structure and natural history of the green turtle (Chelonia mydas) in terms of matriarchal phylogeny. Evolution 46:865–881
Bowen BW, Abreu-Grobois FA, Balazs GH, Kamezaki N, Limpus CJ, Ferl RJ (1995) Trans-Pacific migrations of the loggerhead turtle (Caretta caretta) demonstrated with mitochondrial DNA markers. Proc Natl Acad Sci USA 92:3731–3734
Bowen BW, Bass AL, Chow SM, Bostrom M, Bjorndal KA, Bolten AB, Okuyama T, Bolker BM, Epperly S, Lacasella E, Shaver D, Dodd M, Hopkins-Murphy SR, Musick JA, Swingle M, Rankin-Baransky K, Teas W, Witzell WN, Dutton PH (2004) Natal homing in juvenile loggerhead turtles (Caretta caretta). Mol Ecol 13:3797–3808
Broderick D, Moritz C, Miller JD, Guinea M, Prince RJ, Limpus CJ (1994) Genetic studies of the hawksbill turtle (Eretmochelys imbricata): evidence for multiple stocks in Australian waters. Pac Conserv Biol 1:123–131
Brower AVZ (1999) Delimitation of phylogenetic species with DNA sequences: a critique of Davis and Nixon’s population aggregation analysis. Syst Biol 48:199–213
Campbell CL (2003) Population assessment and management needs of a green turtle, Chelonia mydas, population in the western Caribbean. PhD dissertation, University of Florida, Gainesville, FL
Carr A, Carr MH, Meylan AB (1978) The ecology and migrations of sea turtles, 7. The West Caribbean green turtle colony. Bull Am Mus Nat Hist 162:1–46
Díaz-Fernández R, Okayama T, Uchiyama T, Carrillo E, Espinosa G, Márquez R, Diez C, Koike H (1999) Genetic sourcing for the hawksbill turtle, Eretmochelys imbricata, in the northern Caribbean region. Chelonian Cons Biol 3:296–300
Encalada SE, Lahanas PN, Bjorndal KA, Bolten AB, Miyamoto MM, Bowen BW (1996) Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: a mitochondrial DNA control region sequence assessment. Molec Ecol 5:473–483
Espinosa López G, Hernández Aguilera G, Jager M, Olavarría Gámez K, Ibarra Martín ME, Masselot M, Deutch J (2000) Genetic identification of a nesting colony of green turtles, Chelonia mydas, from the western Cuban shelf. In: Kalb HJ, Wibbels T (eds) Proceedings of the Nineteenth Annual Symposium on Sea Turtle Biology and Conservation. NOAA Technical Memorandum NMFS-SEFSC-443, Miami FL, pp 121–123
FitzSimmons N, Moritz C, Bowen BW (1999) Population identification. In Eckert KL, Bjorndal KA, Abreu Grobois FA, Donnelly M (eds) Management and research techniques for the conservation of sea turtles. IUCN, Gland, Switzerland, pp 72–79
Hatase H, Kinoshita M, Bando T, Kamezaki N, Sato K, Matsuzawa Y, Goto K, Omuta K, Nakashima Y, Takeshita H, SakamotoW (2002) Population structure of loggerhead turtles, Caretta caretta, nesting in Japan: bottlenecks on the Pacific population. Mar Biol 141:299–305
Hillis D, Moritz C, Mable BK (1996) Molecular systematics (2nd edn). Sinauer Associates, Sunderland, MA
Innis MA, Gelfand DH, Sninsky JJ, White TJ (1990) PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA
Lahanas PN, Miyamoto MM, Bjorndal KA, Bolten AB (1994) Molecular evolution and population genetics of Greater Caribbean green turtles (Chelonia mydas) as inferred from mitochondrial DNA control region sequences. Genetica 94:57–67
Lahanas PN, Bjorndal KA, Bolten AB, Encalada SE, Miyamoto MM, Valverde RA, Bowen BW (1998) Genetic composition of a green turtle (Chelonia mydas) feeding ground population: evidence for multiple origins. Mar Biol 130:345–352
Laurent L, Casale P, Bradai MN, Godley BJ, Gerosa G, Broderick AC, Schroth W, Schierwater B, Levy AM, Freggi D, Abd El-Mawla EM, Hadoud DA, El-Gomati H, Domingo M, Hadjichristophorou M, Kornaraki L, Demirayak F, Gautier C (1998) Molecular resolution of marine turtle stock composition in fishery bycatch: a case study in the Mediterranean. Mol Ecol 7:1529–1542
Luke K, Horrocks JA, LeRoux RA, Dutton PH (2004) Origins of green turtle (Chelonia mydas) feeding aggregations around Barbados, West Indies. Mar Biol 144:799–805
Miller JD (1997) Reproduction in sea turtles. In: Lutz PL, Musick JA (eds) The biology of sea turtles. CRC Press, Boca Raton, FL, pp 51–81
Moore MK, Ball Jr RM (2002) Multiple paternity in loggerhead turtle (Caretta caretta) nests on Melbourne Beach, Florida: a microsatellite analysis. Mol Ecol 11:281–288
Mullis KB, Faloona F (1987) Specific synthesis of DNA in vitro via polymerase-catalyzed chain reaction. Methods Enzymol 155:335–350
Norman JA, Moritz C, Limpus CJ (1994) Mitochondrial DNA control region polymorphisms: genetic markers for ecological studies of marine turtles. Molec Ecol 3:363–373
Okuyama T, Bolker BM (2005) Combining genetic and ecological data to estimate sea turtle origins. Ecol Appl 15:315–325
Pearce AF (2001) Contrasting population structure of the loggerhead turtle (Caretta caretta) using mitochondrial and nuclear DNA markers. Masters Thesis, University of Florida, Gainesville, FL
Peare T, Parker PG (1996) Local genetic structure within two rookeries of Chelonia mydas (the green turtle). Heredity 77:619–628
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver 2.000: a software for population genetics data analysis. University of Geneva, Geneva, Switzerland
Schroth W, Streit B, Schierwater B (1996) Evolutionary handicap for turtles. Nature 384:521–522
Seminoff JA (2002) 2002 IUCN red list global status assessment: green turtle (Chelonia mydas). IUCN/SSC Marine Turtle Specialist Group, Gland, Switzerland
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Solow AR, Bjorndal KA, Bolten AB (2002) Annual variation in nesting numbers of marine turtles: the effect of sea surface temperature on re-migration intervals. Ecol Lett 5:742–746
Tamura K, Nei M (1993) Estimation of the number of substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molec Biol Evol 10:512–526
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tiwari M, Bjorndal KA, Bolten AB, Bolker BM (in press) Intraspecific application of the mid-domain effect model: spatial and temporal nest distributions of green turtles, Chelonia mydas, at Tortuguero, Costa Rica. Ecol Lett
Troëng S, Rankin E (2005) Long-term conservation efforts contribute to positive green turtle Chelonia mydas nesting trend at Tortuguero, Costa Rica. Biol Cons 121:111–116
Zaykin DV, Pudovkin AI (1993) Two programs to estimate chi-square values using pseudo-probability test. J Hered 84:152
Acknowledgements
We are grateful to the staff and volunteers of the Caribbean Conservation Corporation who collected the tissue samples at Tortuguero, Costa Rica, in particular Emma Harrison and Catalina Reyes who supervised sample collection in the field, and to Ginger Clark, director of the Genetics Analysis Lab at the University of Florida, who oversaw the determination of genetic sequences. We thank Brian Bowen for constructive comments on the manuscript; Peter Eliazar for assistance with data management; and Charles Baer, Rebecca Kimball, and Michael Miyamoto for advice on genetic analyses. This study was funded by the Disney Wildlife Conservation Fund, the Caribbean Conservation Corporation, and the University of Florida. The Ministry of Environment and Energy of Costa Rica kindly provided collection and CITES export permits. All necessary permits for collection, export and import of samples were obtained. All work was conducted in compliance with the Institutional Animal Care and Use Committee, University of Florida.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P.W. Sammarco, Chauvin
Rights and permissions
About this article
Cite this article
Bjorndal, K.A., Bolten, A.B. & Troëng, S. Population structure and genetic diversity in green turtles nesting at Tortuguero, Costa Rica, based on mitochondrial DNA control region sequences. Marine Biology 147, 1449–1457 (2005). https://doi.org/10.1007/s00227-005-0045-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-005-0045-y