Abstract
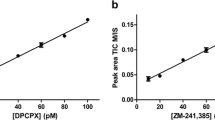
Mass spectrometry (MS) binding assays are a label-free alternative to radioligand or fluorescence binding assays, so the readout is based on direct mass spectrometric detection of the test ligand. The study presented here describes the development and validation of a highly sensitive, rapid, and robust MS binding assay for the quantification of the binding of the metabotropic glutamate 5 (mGlu5) negative allosteric modulator (NAM), MPEP (2-methyl-6-phenylethynylpyridine) at the mGlu5 allosteric binding site. The LC-ESI-MS/MS (liquid chromatography-electrospray ionization-tandem mass spectrometric) analytical method was established and validated with a deuterated analogue of MPEP as an internal standard. The developed MS binding assay described here allowed for the determination of MS binding affinity estimates that were in agreement with affinity estimates obtained from a tritiated MPEP radioligand saturation binding assay, indicating the suitability of this methodology for determining affinity estimates for compounds that target mGlu5 allosteric binding sites.

Graphical abstract






Similar content being viewed by others
References
Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–9.
Hoffmann C, Castro M, Rinken A, Leurs R, Hill SJ, Vischer HF. Ligand residence time at G-protein-coupled receptors - why we should take our time to study it. Mol Pharmacol. 2015;88:552–60.
Swinney DC. Applications of binding kinetics to drug discovery. Pharmaceut Med. 2008;22:23–34.
Lu H, Tonge PJ. Drug-target residence time: critical information for lead optimization. Curr Opin Chem Biol. 2010;14:467–74.
Guo D, Hillger JM, IJzerman AP, Heitman LH. Drug-target residence time - a case for G protein-coupled receptors. Med Res Rev. 2014;34:856–92.
Lefkowitz RJ, Roth J, Pastan I. Radioreceptor assay of adrenocorticotropic hormone: new approach to assay of polypeptide hormones in plasma. Science. 1970;170:633–5.
Pert CB, Snyder SH. Opiate receptor demonstration in nervous tissue. Science. 1973;179:1011–3.
Sridharan R, Zuber J, Connelly SM, Mathew E, Dumont ME. Fluorescent approaches for understanding interactions of ligands with G protein coupled receptors. Biochim Biophys Acta. 2014;1838:15–33.
Hovius R, Vallotton P, Wohland T, Vogel H. Fluorescence techniques: shedding light on ligand-receptor interactions. Trends Pharmacol Sci. 2000;21:266–73.
Zwier JM, Roux T, Cottet M, Durroux T, Douzon S, Bdioui S, et al. A fluorescent ligand-binding alternative using Tag-lite® technology. J Biomol Screen. 2010;15:1248–59.
Zepperitz C, Höfner G, Wanner KT. MS-binding assays: kinetic, saturation, and competitive experiments based on quantitation of bound marker as exemplified by the GABA transporter mGAT1. ChemMedChem. 2006;1:208–17.
Hess M, Höfner G, Wanner KT. (S)- and (R)-fluoxetine as native markers in mass spectrometry (MS) binding assays addressing the serotonin transporter. ChemMedChem. 2011;6:1900–8.
Hess M, Höfner G, Wanner KT. Development and validation of a rapid LC-ESI-MS/MS method for quantification of fluoxetine and its application to MS binding assays. Anal Bioanal Chem. 2011;400:3505–15.
Schmitt S, Höfner G, Wanner KT. MS transport assays for γ-aminobutyric acid transporters an efficient alternative for radiometric assays. Anal Chem. 2014;86:7575–83.
Grimm SH, Höfner G, Wanner KT. Development and validation of an LC-ESI-MS/MS method for the triple reuptake inhibitor indatraline enabling its quantification in MS binding assays. Anal Bioanal Chem. 2015;407:471–85.
Grimm SH, Höfner G, Wanner KT. MS binding assays for the three monoamine transporters using the triple reuptake inhibitor (1R,3S)-indatraline as native marker. ChemMedChem. 2015;10:1027–39.
Schuller M, Höfner G, Wanner KT. Simultaneous multiple MS binding assays addressing D1 and D2 dopamine receptors. Chem Med Chem. 2017;12:1–11.
Sichler S, Höfner G, Rappenglück S, Wein T, Niessenb KV, Seeger T, et al. Development of MS binding assays targeting the binding site of MB327 at the nicotinic acetylcholine receptor. Toxicol Lett. 2018;293:172–83.
Neiens P, De Simone A, Ramershoven A, Höfner G, Allmendinger L, Wanner KT. Development and validation of an LC-ESI-MS/MS method for the quantification of D-84, reboxetine, and citalopram for their use in MS binding assays addressing the monoamine transporters hDAT, hSERT, and hNET. Biomed Chromatogr. 2018;32:e4231.
Neiens P, De Simone A, Höfner G, Wanner KT. Simultaneous multiple MS binding assays for the dopamine, norepinephrine, and serotonin transporters. ChemMedChem. 2018;13:453–63.
Massink A, Holzheimer M, Hölscher A, Louvel J, Guo D, Spijksma G, et al. Mass spectrometry-based ligand binding assays on adenosine A1 and A2A receptors. Purinergic Signal. 2015;11:581–94.
Rossato M, Miralles G, M’Kadmi C, Maingot M, Amblard M, Mouillac B, et al. Quantitative MALDI-MS binding assays: an alternative to radiolabeling. ChemMedChem. 2016;11:2582–7.
Cheignon C, Cordeau E, Prache N, Cantel S, Martinez J, Subra G, et al. Receptor-ligand interaction measured by inductively coupled plasma mass spectrometry and selenium labeling. J Med Chem. 2018;61:10173–84.
Nasrallah C, Rottier K, Marcellin R, Compan V, Font J, Llebaria A, et al. Direct coupling of detergent purified human mGlu5 receptor to the heterotrimeric G proteins Gq and Gs. Sci Rep Nature. 2018;8:4407.
Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322.
Nicoletti F, Bockaert J, Collingridge GL, Conn PJ, Ferraguti F, Schoepp DD, et al. Metabotropic glutamate receptors: from the workbench to the bedside. Neuropharmacology. 2011;60:1017–41.
Pereira V, Goudet C. Emerging trends in pain modulation by metabotropic glutamate receptors. Front Mol Neurosci. 2018;11:464.
Walker AG, Conn PJ. Group I and group II metabotropic glutamate receptor allosteric modulators as novel potential antipsychotics. Curr Opin Pharmacol. 2015;20:40–5.
Marszalek-Grabska M, Gibula-Bruzda E, Bodzon-Kulakowaka A, Suder P, Gawel K, Talarek S, et al. ADX-47273, a mGlu5 receptor positive allosteric modulator, attenuates deficits in cognitive flexibility induced by withdrawal from ‘binge-like’ ethanol exposure in rats. Behav Brain Res. 2018;338:9–16.
Felts AS, Bollinger KA, Brassard CJ, Rodriguez AL, Morrison RD, Scott DJ, et al. Discovery of 4-alkoxy-6-methylpicolinamide negative allosteric modulators of metabotropic glutamate receptor subtype 5. Bioorg Med Chem Lett. 2019;29:47–50.
Cavallone LF, Montana MC, Frey K, Kallogjeri D, Wages JM, Rodebaugh TL, et al. The metabotropic glutamate receptor 5 negative allosteric modulator fenobam: pharmacokinetics, side effects, and analgesic effects in healthy human subjects. Pain. 2020;161:135–46.
Gasparini F, Lingenhöhl K, Stoehr N, Flor PJ, Heinrich M, Vranesic I, et al. 2-Methyl-6-(phenylethynyl)-pyridine (MPEP), a potent, selective and systemically active mGlu5 receptor antagonist. Neuropharmacology. 1999;38:1493–503.
Nagel J, Greco S, Parsons CG, Flik G, Tober C, Klein KU, et al. Brain concentrations of mGluR5 negative allosteric modulator MTEP in relation to receptor occupancy - comparison to MPEP. Pharmacol Rep. 2015;67:624–30.
Felts AS, Rodriguez AL, Morrison RD, Venable DF, Manka JT, Bates BS, et al. Discovery of VU0409106: a negative allosteric modulator of mGlu5 with activity in a mouse model of anxiety. Bioorg Med Chem Lett. 2013;23:5779–85.
Alagille D, Baldwin RM, Roth BL, Wroblewski JT, Grajkowska E, Tamagnan GD. Synthesis and receptor assay of aromatic-ethynyl-aromatic derivatives with potent mGluR5 antagonist activity. Bioorg Med Chem. 2005;13:197–209.
Xu Z, Lou Y, Chen L, Zeng K, Sun Q, Lei X WO 2019144765.
Porter RH, Jaeschke G, Spooren W, Ballard TM, Büttelmann B, Kolczewski S, et al. Fenobam: a clinically validated nonbenzodiazepine anxiolytic is a potent, selective, and noncompetitive mGlu5 receptor antagonist with inverse agonist activity. J Pharmacol Exp Ther. 2005;315:711–21.
US Department of Health and Human Services Food and Drug Administration. Guidance for industry: bioanalytical method validation, https://www.fda.gov/media/70858/download.
Acknowledgments
We thank Prof. Jean-Philippe Pin (IGF, University of Montpellier, CNRS, INSERM, Montpellier) for scientific support and contributions to this work.
Funding
This work was financially supported by FEDER/Ministerio de Ciencia, Innovación y Universidades - Agencia Estatal de Investigación (CTQ2017-89222-R and PCI2018-093047), by the Catalan government (2017SGR1604), by Neuron-ERANET, by the Agence Nationale de la Recherche (ANR-17-NEU3-0001 under the frame of Neuron Cofund) and by the Programme International de Collaboration Scientifique (PICS 08212) of the CNRS. AEB was supported by the Labex EpiGenMed (program « Investissements d’avenir », ANR-10-LABX-12-01).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 340 kb)
Rights and permissions
About this article
Cite this article
Ricart-Ortega, M., Berizzi, A.E., Catena, J. et al. Development and validation of a mass spectrometry binding assay for mGlu5 receptor. Anal Bioanal Chem 412, 5525–5535 (2020). https://doi.org/10.1007/s00216-020-02772-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02772-9