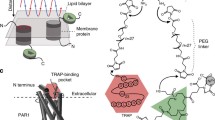
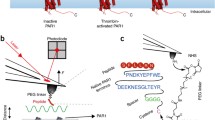
Abstract
Cell surface receptors, often called transmembrane receptors, are key cellular components as they control and mediate cell communication and signalling, converting extracellular signals into intracellular signals. Elucidating the molecular details of ligand binding (cytokine, growth factors, hormones, pathogens,...) to cell surface receptors and how this binding triggers conformational changes that initiate intracellular signalling is needed to improve our understanding of cellular processes and for rational drug design. Unfortunately, the molecular complexity and high hydrophobicity of membrane proteins significantly hamper their structural and functional characterization in conditions mimicking their native environment. With its piconewton force sensitivity and (sub)nanometer spatial resolution, together with the capability of operating in liquid environment and at physiological temperature, atomic force microscopy (AFM) has proven to be one of the most powerful tools to image and quantify receptor-ligand bonds in situ under physiologically relevant conditions. In this article, a brief overview of the rapid evolution of AFM towards quantitative biological mapping will be given, followed by selected examples highlighting the main advances that AFM-based ligand-receptor studies have brought to the fields of cell biology, immunology, microbiology, and virology, along with future prospects and challenges.

Graphical abstract




Similar content being viewed by others
References
Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56(9):930–3. https://doi.org/10.1103/PhysRevLett.56.930.
Pavliček N, Gross L. Generation, manipulation and characterization of molecules by atomic force microscopy. Nat Rev Chem. 2017;1:0005. https://doi.org/10.1038/s41570-016-0005.
Alsteens D, Gaub HE, Newton R, Pfreundschuh M, Gerber C, Müller DJ. Atomic force microscopy-based characterization and design of biointerfaces. Nat Rev Mat. 2017;2(5). doi:https://doi.org/10.1038/natrevmats.2017.8.
Frisbie CD, Rozsnyai LF, Noy A, Wrighton MS, Lieber CM. Functional group imaging by chemical force microscopy. Science (New York, NY). 1994;265(5181):2071–4. https://doi.org/10.1126/science.265.5181.2071.
Ludwig M, Dettmann W, Gaub HE. Atomic force microscope imaging contrast based on molecular recognition. Biophys J. 1997;72(1):445–8.
Lee GU, Kidwell DA, Colton RJ. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir. 1994;10(2):354–7. https://doi.org/10.1021/la00014a003.
Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science (New York, NY). 1994;264(5157):415–7. https://doi.org/10.1126/science.8153628.
Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc Natl Acad Sci U S A. 1996;93(8):3477–81. https://doi.org/10.1073/pnas.93.8.3477.
Grandbois M, Dettmann W, Benoit M, Gaub HE. Affinity imaging of red blood cells using an atomic force microscope. J Histochem Cytochem. 2000;48(5):719–24. https://doi.org/10.1177/002215540004800516.
Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat Cell Biol. 2000;2(6):313–7. https://doi.org/10.1038/35014000.
Alsteens D, Beaussart A, Derclaye S, El-Kirat-Chatel S, Park HR, Lipke PN, et al. Single-cell force spectroscopy of Als-mediated fungal adhesion. Anal Methods. 2013;5(15):3657–62. https://doi.org/10.1039/C3AY40473K.
Dufrêne YF, Ando T, Garcia R, Alsteens D, Martinez-Martin D, Engel A, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12(4):295–307. https://doi.org/10.1038/nnano.2017.45.
Alsteens D, Trabelsi H, Soumillion P, Dufrêne YF. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat Commun. 2013;4:2926. https://doi.org/10.1038/ncomms3926.
Laskowski PR, Pfreundschuh M, Stauffer M, Ucurum Z, Fotiadis D, Muller DJ. High-resolution imaging and multiparametric characterization of native membranes by combining confocal microscopy and an atomic force microscopy-based toolbox. ACS Nano. 2017;11(8):8292–301. https://doi.org/10.1021/acsnano.7b03456.
Pfreundschuh M, Harder D, Ucurum Z, Fotiadis D, Muller DJ. Detecting ligand-binding events and free energy landscape while imaging membrane receptors at subnanometer resolution. Nano Lett. 2017;17(5):3261–9. https://doi.org/10.1021/acs.nanolett.7b00941.
Fuhrmann A, Ros R. Single-molecule force spectroscopy: a method for quantitative analysis of ligand-receptor interactions. Nanomedicine (London, England). 2010;5(4):657–66. https://doi.org/10.2217/nnm.10.26.
Wildling L, Rankl C, Haselgrubler T, Gruber HJ, Holy M, Newman AH, et al. Probing binding pocket of serotonin transporter by single molecular force spectroscopy on living cells. J Biol Chem. 2012;287(1):105–13. https://doi.org/10.1074/jbc.M111.304873.
Chtcheglova LA, Hinterdorfer P. Simultaneous AFM topography and recognition imaging at the plasma membrane of mammalian cells. Semin Cell Dev Biol. 2018;73:45–56. https://doi.org/10.1016/j.semcdb.2017.08.025.
Koehler M, Macher G, Rupprecht A, Zhu R, Gruber HJ, Pohl EE, et al. Combined recognition imaging and force spectroscopy: a new mode for mapping and studying interaction sites at low lateral density. Sci Adv. 2017;9(1):128–34. https://doi.org/10.1166/sam.2017.3066.
Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356. https://doi.org/10.1038/nature08144.
Alsteens D, Pfreundschuh M, Zhang C, Spoerri PM, Coughlin SR, Kobilka BK, et al. Imaging G protein–coupled receptors while quantifying their ligand-binding free-energy landscape. Nat Methods. 2015;12:845. https://doi.org/10.1038/nmeth.3479.
Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397(6714):50–3. https://doi.org/10.1038/16219.
Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci U S A. 2012;109(34):13573–8. https://doi.org/10.1073/pnas.1202946109.
Pfreundschuh M, Alsteens D, Wieneke R, Zhang C, Coughlin SR, Tampé R, et al. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat Commun. 2015;6:8857. https://doi.org/10.1038/ncomms9857.
Knoops B, Becker S, Poncin MA, Glibert J, Derclaye S, Clippe A, Alsteens D. Specific interactions measured by AFM on living cells between peroxiredoxin-5 and TLR4: relevance for mechanisms of innate immunity. Cell Chem Biol 2018;25(5):550–559 e3. doi:https://doi.org/10.1016/j.chembiol.2018.02.006.
Dufrêne YF. Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. mBio. 2014;5(4):e01363–14. https://doi.org/10.1128/mBio.01363-14.
Mitchell G, Lamontagne CA, Brouillette E, Grondin G, Talbot BG, Grandbois M, et al. Staphylococcus aureus SigB activity promotes a strong fibronectin-bacterium interaction which may sustain host tissue colonization by small-colony variants isolated from cystic fibrosis patients. Mol Microbiol. 2008;70(6):1540–55. https://doi.org/10.1111/j.1365-2958.2008.06511.x.
Buck AW, Fowler VG, Yongsunthon R, Liu J, DiBartola AC, Que Y-A, et al. Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir. 2010;26(13):10764–70. https://doi.org/10.1021/la100549u.
Casillas-Ituarte NN, Lower BH, Lamlertthon S, Fowler VG Jr, Lower SK. Dissociation rate constants of human fibronectin binding to fibronectin-binding proteins on living Staphylococcus aureus isolated from clinical patients. J Biol Chem. 2012;287(9):6693–701. https://doi.org/10.1074/jbc.M111.285692.
Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci U S A. 2011;108(45):18372–7. https://doi.org/10.1073/pnas.1109071108.
Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115(2):217–28.
Herman P, El-Kirat-Chatel S, Beaussart A, Geoghegan JA, Foster TJ, Dufrêne YF. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol Microbiol. 2014;93(2):356–68. https://doi.org/10.1111/mmi.12663.
Herman-Bausier P, Labate C, Towell AM, Derclaye S, Geoghegan JA, Dufrêne YF. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc Natl Acad Sci U S A. 2018;115(21):5564–9. https://doi.org/10.1073/pnas.1718104115.
Vitry P, Valotteau C, Feuillie C, Bernard S, Alsteens D, Geoghegan JA, Dufrêne YF. Force-induced strengthening of the interaction between Staphylococcus aureus clumping factor B and loricrin. MBio. 2017;8(6). https://doi.org/10.1128/mBio.01748-17.
Milles LF, Schulten K, Gaub HE, Bernardi RC. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science (New York, NY). 2018;359(6383):1527–33. https://doi.org/10.1126/science.aar2094.
Milles LF, Unterauer EM, Nicolaus T, Gaub HE. Calcium stabilizes the strongest protein fold. Nat Commun. 2018;9(1):4764. https://doi.org/10.1038/s41467-018-07145-6.
Viela F, Prystopiuk V, Leprince A, Mahillon J, Speziale P, Pietrocola G, Dufrêne YF. Binding of Staphylococcus aureus protein A to von Willebrand factor is regulated by mechanical force. MBio. 2019;10(2). https://doi.org/10.1128/mBio.00555-19.
Alsteens D, Dupres V, Yunus S, Latgé J-P, Heinisch JJ, Dufrêne YF. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir. 2012;28(49):16738–44. https://doi.org/10.1021/la303891j.
Chang MI, Panorchan P, Dobrowsky TM, Tseng Y, Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J Virol. 2005;79(23):14748–55. https://doi.org/10.1128/jvi.79.23.14748-14755.2005.
Dobrowsky TM, Zhou Y, Sun SX, Siliciano RF, Wirtz D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. J Virol. 2008;82(14):7022–33. https://doi.org/10.1128/jvi.00053-08.
Rankl C, Kienberger F, Wildling L, Wruss J, Gruber HJ, Blaas D, et al. Multiple receptors involved in human rhinovirus attachment to live cells. Proc Natl Acad Sci U S A. 2008;105(46):17778–83. https://doi.org/10.1073/pnas.0806451105.
Sieben C, Kappel C, Zhu R, Wozniak A, Rankl C, Hinterdorfer P, et al. Influenza virus binds its host cell using multiple dynamic interactions. Proc Natl Acad Sci U S A. 2012;109(34):13626–31. https://doi.org/10.1073/pnas.1120265109.
Delguste M, Peerboom N, Le Brun G, Trybala E, Olofsson S, Bergström T, et al. Regulatory mechanisms of the mucin-like region on herpes simplex virus during cellular attachment. ACS Chem Biol. 2019;14(3):534–42. https://doi.org/10.1021/acschembio.9b00064.
Alsteens D, Newton R, Schubert R, Martinez-Martin D, Delguste M, Roska B, et al. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat Nanotechnol. 2017;12(2):177–83. https://doi.org/10.1038/nnano.2016.228.
Newton R, Delguste M, Koehler M, Dumitru AC, Laskowski PR, Muller DJ, et al. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat Protoc. 2017;12(11):2275–92. https://doi.org/10.1038/nprot.2017.112.
Delguste M, Zeippen C, Machiels B, Mast J, Gillet L, Alsteens D. Multivalent binding of herpesvirus to living cells is tightly regulated during infection. Sci Adv. 2018;4(8):eaat1273. https://doi.org/10.1126/sciadv.aat1273.
Yu H, Siewny MGW, Edwards DT, Sanders AW, Perkins TT. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science (New York, NY). 2017;355(6328):945–50. https://doi.org/10.1126/science.aah7124.
Harke B, Chacko JV, Haschke H, Canale C, Diaspro A. A novel nanoscopic tool by combining AFM with STED microscopy. Opt Nanoscopy. 2012;1(1):3. https://doi.org/10.1186/2192-2853-1-3.
Chacko JV, Zanacchi FC, Diaspro A. Probing cytoskeletal structures by coupling optical superresolution and AFM techniques for a correlative approach. Cytoskeleton (Hoboken, NJ). 2013;70(11):729–40. https://doi.org/10.1002/cm.21139.
Curry N, Ghézali G, Kaminski Schierle GS, Rouach N, Kaminski CF. Correlative STED and Atomic Force Microscopy on Live Astrocytes Reveals Plasticity of Cytoskeletal Structure and Membrane Physical Properties during Polarized Migration. Front Cell Neurosci. 2017;11:104. https://doi.org/10.3389/fncel.2017.00104.
Funding
This work was supported by the Fonds National de la Recherche Scientifique (F.R.S.-FNRS grant number: PDR T.0090.15 to D.A.), the Research Department of the Communauté française de Belgique (Concerted Research Action), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the ‘MOVE-IN Louvain’ Incoming post-doc Fellowship programme, and the European Molecular Biology Organization (EMBO ALTF 542-2018 to C.L.). D.A. is Research Associate at the FNRS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lo Giudice, C., Dumitru, A.C. & Alsteens, D. Probing ligand-receptor bonds in physiologically relevant conditions using AFM. Anal Bioanal Chem 411, 6549–6559 (2019). https://doi.org/10.1007/s00216-019-02077-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-02077-6