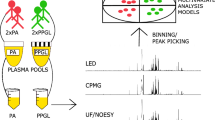
Abstract
Differential mobility spectrometry (DMS) has been gaining popularity in small molecule analysis over the last few years due to its selectivity towards a variety of isomeric compounds. While DMS has been utilized in targeted liquid chromatography-mass spectrometry (LC-MS), its use in untargeted discovery workflows has not been systematically explored. In this contribution, we propose a novel workflow for untargeted metabolomics based solely on DMS separation in a clinically relevant chronic kidney disease (CKD) patient population. We analyzed ten plasma samples from early- and late-stage CKD patients. Peak finding, alignment, and filtering steps performed on the DMS-MS data yielded a list of 881 metabolic features (unique mass-to-charge and migration time combinations). Differential analysis by CKD patient group revealed three main features of interest. One of them was putatively identified as bilirubin based on high-accuracy MS data and comparison of its optimum compensation voltage (COV) with that of an authentic standard. The DMS-MS analysis was four times faster than a typical HPLC-MS run, which suggests a potential for the utilization of this technique in screening studies. However, its lower separation efficiency and reduced signal intensity make it less suitable for low-abundant features. Fewer features were detected by the DMS-based platform compared with an HPLC-MS-based approach, but importantly, the two approaches resulted in different features. This indicates a high degree of orthogonality between HPLC- and DMS-based approaches and demonstrates the need for larger studies comparing the two techniques. The workflow described here can be adapted for other areas of metabolomics and has a value as a prescreening method to develop semi-targeted workflows and as a faster alternative to HPLC in large biomedical studies.






Similar content being viewed by others
Abbreviations
- ACN:
-
Acetonitrile
- BMI:
-
Body mass index
- CKD:
-
Chronic kidney disease
- COV:
-
Compensation voltage
- CPROBE:
-
Clinical Phenotyping Resource and Biobank Core at the University of Michigan
- cps:
-
Counts per second
- DMS:
-
Differential mobility spectrometry
- eGFR:
-
Estimated glomerular filtration rate
- FC:
-
Fold change
- FDR:
-
False discovery rate
- FOI:
-
Feature of interest
- HPLC:
-
High-performance liquid chromatography
- iPrOH:
-
Isopropanol
- MS:
-
Mass spectrometry
- PCA:
-
Principal component analysis
- PLS-DA:
-
Partial least squares-discriminant analysis
- PP:
-
Pooled plasma
- RP:
-
Reversed-phase
- (R)SD:
-
(Relative) standard deviation
- SV:
-
Separation voltage
- TIC:
-
Total ion current
- UPCR:
-
Urinary protein-to-creatinine ratio
- VIP:
-
Variable importance in projection (PLS-DA)
- XIC:
-
Extracted ion chromatogram
References
Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448.
Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123(10):4309–17.
Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8(1):615.
Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 2016;1(15).
Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85.
Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. New Engl J Med. 2013;368(17):1575–84.
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57.
Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res. 2011;71(24):7376–86.
Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, et al. Impaired β-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J Am Soc Nephrol. 2018;29(1):295–306.
Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85(5):1214–24.
Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, et al. Lipidomic signature of progression of chronic kidney disease in the chronic renal insufficiency cohort. Kidney Int Reports. 2016;1(4):256–68.
Hocher B, Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol. 2017;13:269.
Campbell JL, Blanc JYL, Kibbey RG. Differential mobility spectrometry: a valuable technology for analyzing challenging biological samples. Bioanalysis. 2015;7(7):853–6.
Liu C, Gómez-Ríos GA, Schneider BB, Le Blanc JCY, Reyes-Garcés N, Arnold DW, et al. Fast quantitation of opioid isomers in human plasma by differential mobility spectrometry/mass spectrometry via SPME/open-port probe sampling interface. Anal Chim Acta. 2017;991:89–94.
Ayodeji I, Vazquez T, Bailey R, Evans-Nguyen T. Rapid pre-filtering of amphetamine and derivatives by direct analysis in real time (DART)-differential mobility spectrometry (DMS). Anal Methods. 2017;9(34):5044–51.
Chen Z, Coy SL, Pannkuk EL, Laiakis EC, Fornace AJ, Vouros P. Differential mobility spectrometry-mass spectrometry (DMS-MS) in radiation biodosimetry: rapid and high-throughput quantitation of multiple radiation biomarkers in nonhuman primate urine. J Am Soc Mass Spectr. 2018.
Wernisch S, Afshinnia F, Rajendiran T, Pennathur S. Probing the application range and selectivity of a differential mobility spectrometry–mass spectrometry platform for metabolomics. Anal Bioanal Chem. 2018;410(12):2865–77.
Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. Ion mobility–mass spectrometry. J Mass Spectrom. 2008;43(1):1–22.
Wernisch S, Afshinnia F, Rajendiran TM, Pennathur S. Differential mobility – mass spectrometry metabolomics platform for biomarker discovery in chronic kidney disease. Annual Meeting of the American Society for Mass Spectrometry; 2017; Indianapolis, IN.
Stevens LA, Schmid CH, Greene T, Zhang YL, Beck GJ, Froissart M, et al. Comparative performance of the CKD Epidemiology Collaboration (CKD-EPI) and the Modification of Diet in Renal Disease (MDRD) Study equations for estimating GFR levels above 60 mL/min/1.73 m2. Am J Kidney Dis. 2010;56(3):486–95.
Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W7.
Brereton RG, Lloyd GR. Partial least squares discriminant analysis: taking the magic away. J Chemom. 2014;28(4):213–25.
Anwar A, Psutka J, Walker SWC, Dieckmann T, Janizewski JS, Larry Campbell J, et al. Separating and probing tautomers of protonated nucleobases using differential mobility spectrometry. Int J Mass Spectrom. 2017.
Acknowledgements
The authors thank Dr. Farsad Afshinnia (University of Michigan) for helping in statistical data analysis and Dr. J. Larry Campbell (Sciex) for providing access to MarkerView software. We also appreciate the assistance of the Michigan Kidney Translational Core Clinical Phenotyping Resource and Biobank Core Investigator Group. It includes Matthias Kretzler and Debbie Gipson (University of Michigan, Ann Arbor), Keith Bellovich (St. Clair Nephrology Research, Detroit), Zeenat Bhat (Wayne State University, Detroit), Crystal Gadegbeku (Temple University Health System, Philadelphia), Susan Massengill (Levin Children’s Hospital, Charlotte), and Kalyani Perumal (JH Stroger Hospital, Chicago).
Funding
This work was supported by the Postdoctoral Translational Science Program from the Michigan Institute for Clinical and Health Research UL1TR000433 (to S.W.) and National Institutes of Health grants P30DK089503, DK082841, P30DK081943, U2C ES026553, and DK097153 (to S.P.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All human studies were approved by the Institutional Review Board (IRB) for the University of Michigan and the CPROBE ancillary studies committee.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Published in the topical collection Close-Up of Current Developments in Ion Mobility Spectrometry with guest editor Gérard Hopfgartner.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 907 kb)
Rights and permissions
About this article
Cite this article
Wernisch, S., Pennathur, S. Application of differential mobility-mass spectrometry for untargeted human plasma metabolomic analysis. Anal Bioanal Chem 411, 6297–6308 (2019). https://doi.org/10.1007/s00216-019-01719-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-01719-z