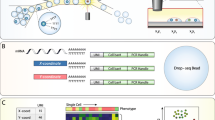
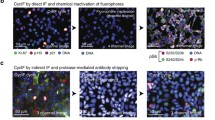
Abstract
The quantity and activity of proteins in many biological systems exhibit prominent heterogeneities. Single-cell analytical methods can resolve subpopulations and dissect their unique signatures from heterogeneous samples, enabling a clarifying view of the biological process. Over the last 5 years, technologies for single-cell protein analysis have significantly advanced. In this article, we highlight a branch of those technology developments involving fluorescence-based approaches, with a focus on the methods that increase the ability to multiplex and enable dynamic measurements. We also analyze the limitations of these techniques and discuss current challenges in the field, with the hope that more transformative platforms can soon emerge.








Similar content being viewed by others
References
Hunter T. Signaling--2000 and beyond. Cell. 2000;1:113–27.
Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;2:295–309.
Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;3:471–84.
Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;6980:332–7.
Vetere A, Choudhary A, Burns SM, Wagner BK. Targeting the pancreatic beta-cell to treat diabetes. Nat Rev Drug Discov. 2014;4:278–89.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;1:57–70.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;5:646–74.
Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;2:85–96.
Rawlings DJ, Metzler G, Wray-Dutra M, Jackson SW. Altered B cell signalling in autoimmunity. Nat Rev Immunol. 2017;7:421–36.
Gross S, Rahal R, Stransky N, Lengauer C, Hoeflich KP. Targeting cancer with kinase inhibitors. J Clin Invest. 2015;5:1780–9.
Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;1:28–37.
Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;7:422–39.
Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, et al. Intratumoural heterogeneity generated by notch signalling promotes small-cell lung cancer. Nature. 2017;7654:360–4.
Welch DR. Tumor heterogeneity—a ‘contemporary concept’ founded on historical insights and predictions. Cancer Res. 2016;1:4–6.
Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;17:738–43.
Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;3:175–88.
Prakadan SM, Shalek AK, Weitz DA. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat Rev Genet. 2017;6:345–61.
Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;4:780–91.
Yu J, Zhou J, Sutherland A, Wei W, Shin YS, Xue M, et al. Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu Rev Anal Chem. 2014;1:275–95.
Su Y, Wei W, Robert L, Xue M, Tsoi J, Garcia-Diaz A, et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci. 2017;52:13679–84.
Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;5:252–62.
Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;1:35–48.
Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–16.
Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci. 2015;7:E607–E15.
Yang L, Wang Z, Deng Y, Li Y, Wei W, Shi Q. Single-cell, multiplexed protein detection of rare tumor cells based on a beads-on-barcode antibody microarray. Anal Chem. 2016;22:11077–83.
Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;6233:aaa6090.
Mondal M, Liao R, Xiao L, Eno T, Guo J. Highly multiplexed single-cell in situ protein analysis with cleavable fluorescent antibodies. Angew Chem Int Ed. 2017;10:2636–9.
Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;5957:1220–4.
Shin YS, Remacle F, Fan R, Hwang K, Wei W, Ahmad H, et al. Protein signaling networks from single cell fluctuations and information theory profiling. Biophys J. 2011;10:2378–86.
Karl FA. Free energy principle for biological systems. Entropy (Basel). 2012;11:2100–21.
Komatsuzaki T, Baba A, Kawai S, Toda M, Straub JE, Berry RS. Ergodic problems for real complex systems in chemical physics. Advancing theory for kinetics and dynamics of complex, many-dimensional systems: clusters and proteins. Adv Chem Phys. 2011;145:171–220.
Ridden SJ, Chang HH, Zygalakis KC, MacArthur BD. Entropy, ergodicity, and stem cell multipotency. Phys Rev Lett. 2015;20:208103.
Weigel AV, Simon B, Tamkun MM, Krapf D. Ergodic and nonergodic processes coexist in the plasma membrane as observed by single-molecule tracking. Proc Natl Acad Sci. 2011;16:6438–43.
Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci. 2012;5:1607–12.
Zotter A, Bäuerle F, Dey D, Kiss V, Schreiber G. Quantifying enzyme activity in living cells. J Biol Chem. 2017;38:15838–48.
Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem Sci. 2017;2:111–29.
Ni Q, Mehta S, Zhang J. Live-cell imaging of cell signaling using genetically encoded fluorescent reporters. FEBS J. 2017;2:203–19.
Conlon P, Gelin-Licht R, Ganesan A, Zhang J, Levchenko A. Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc Natl Acad Sci. 2016;40:E5896–E905.
Mehta S, Zhang Y, Roth RH, Zhang J-f, Mo A, Tenner B, et al. Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol. 2018;10:1215–25.
Gross SM, Dane MA, Bucher E, Heiser LM. High resolution AKT signaling in individual cells. bioRxiv. 2018. https://doi.org/10.1101/373993
Shao S, Li Z, Cheng H, Wang S, Perkins NG, Sarkar P, et al. Chemical approach for profiling intracellular AKT signaling dynamics from single cells. J Am Chem Soc. 2018;42:13586–9.
Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;1:83.
Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84.
Buescher JM, Driggers EM. Integration of omics: more than the sum of its parts. Cancer Metab. 2016;4:4.
Acknowledgements
We thank Prof. Jin Zhang for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Published in the topical collection Young Investigators in (Bio-)Analytical Chemistry with guest editors Erin Baker, Kerstin Leopold, Francesco Ricci, and Wei Wang.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, S., Ji, F., Li, Z. et al. Fluorescence imaging-based methods for single-cell protein analysis. Anal Bioanal Chem 411, 4339–4347 (2019). https://doi.org/10.1007/s00216-019-01694-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-01694-5