Abstract
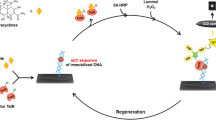
Analytical microarrays feature great capabilities for simultaneous detection and quantification of multiple analytes in a single measurement. In this work, we present a rapid and simple method for bulk preparation of microarrays on polycarbonate sheets. Succinylated Jeffamine® ED-2003 was screen printed on polycarbonate sheets to create a polyfunctional shielding layer by baking at 100 °C. After microdispension of capture probes (antibodies, oligonucleotides, or small molecules) in a microarray format, chips were assembled with a flow cell from double-sided tape. It was shown that the shielding layer was firmly coated and suppressed unspecific binding of proteins. Universal applicability was demonstrated by transferring established flow-based chemiluminescence microarray measurement principles from glass slides to polycarbonate chips without loss of analytical performance. Higher chemiluminescence signals could be generated by performing heterogeneous asymmetric recombinase polymerase amplification on polycarbonate chips. Similar results could be shown for sandwich microarray immunoassays. Beyond that, lower inter- and intra-assay variances could be measured for the analysis of Legionella pneumophila Serogroup 1, strain Bellingham-1. Even surface regeneration of indirect competitive immunoassays was possible, achieving a limit of detection of 0.35 ng L−1 for enrofloxacin with polycarbonate microarray chips. Succinylated Jeffamine ED-2003 coated polycarbonate chips have great potential to replace microtiter plates by flow-based chemiluminescence microarrays for rapid analysis. Therefore, it helps analytical microarrays to advance into routine analysis and diagnostics.

ᅟ








Similar content being viewed by others
References
Kober C, Niessner R, Seidel M. Quantification of viable and non-viable Legionella spp. by heterogeneous asymmetric recombinase polymerase amplification (haRPA) on a flow-based chemiluminescence microarray. Biosens Bioelectron. 2018;100:49–55.
Seidel M, Niessner R. Chemiluminescence microarrays in analytical chemistry: a critical review. Anal Bioanal Chem. 2014;406(23):5589–612.
Kloth K, Rye-Johnsen M, Didier A, Dietrich R, Märtlbauer E, Niessner R, et al. A regenerable immunochip for the rapid determination of 13 different antibiotics in raw milk. Analyst. 2009;134(7):1433–9.
Sola L, Álvarez J, Cretich M, Swann MJ, Chiari M, Hill D. Characterization of porous alumina membranes for efficient, real-time, flow through biosensing. J Membr Sci. 2015;476:128–35.
Bañuls MJ, Morais SB, Tortajada-Genaro LA, Maquieira A. Microarray developed on plastic substrates. In: Li PC, Sedighi A, Wang L, editors. Microarray Technology. Berlin: Springer; 2015.
Besmer MD, Epting J, Page RM, Sigrist JA, Huggenberger P, Hammes F. Online flow cytometry reveals microbial dynamics influenced by concurrent natural and operational events in groundwater used for drinking water treatment. Sci Rep. 2016;6:38462–72.
Grover D, Zhang Z, Readman J, Zhou J. A comparison of three analytical techniques for the measurement of steroidal estrogens in environmental water samples. Talanta. 2009;78(3):1204–10.
Smart AS, Tingley R, Weeks AR, van Rooyen AR, McCarthy MA. Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol Appl. 2015;25(7):1944–52.
Longwell CK, Labanieh L, Cochran JR. High-throughput screening technologies for enzyme engineering. Curr Opin Biotechnol. 2017;48:196–202.
Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–9.
Gao H, Liu M, Zhou X, Liu J, Zhuo Y, Gou Z, et al. Identification of avermectin-high-producing strains by high-throughput screening methods. Appl Microbiol Biotechnol. 2010;85(4):1219–25.
Xu ZN, Shen WH, Chen XY, Lin JP, Cen PL. A high-throughput method for screening of rapamycin-producing strains of Streptomyces hygroscopicus by cultivation in 96-well microtiter plates. Biotechnol Lett. 2005;27(15):1135–40.
Tronser T, Popova AA, Levkin PA. Miniaturized platform for high-throughput screening of stem cells. Curr Opin Biotechnol. 2017;46:141–9.
Gong P, Grainger DW, Surfaces N. In: Rampal JB, editor. Microarrays: volume 1: synthesis methods. Totowa, NJ: Humana Press; 2007. p. 59–92.
Cretich M, Damin F, Chiari M. Protein microarray technology: how far off is routine diagnostics? Analyst. 2013;139(3):528–42.
Wolter A, Niessner R, Seidel M. Preparation and characterization of functional poly (ethylene glycol) surfaces for the use of antibody microarrays. Anal Chem. 2007;79(12):4529–37.
Mehne J, Markovic G, Pröll F, Schweizer N, Zorn S, Schreiber F, et al. Characterisation of morphology of self-assembled PEG monolayers: a comparison of mixed and pure coatings optimised for biosensor applications. Anal Bioanal Chem. 2008;391(5):1783–91.
Piehler J, Brecht A, Valiokas R, Liedberg B, Gauglitz G. A high-density poly (ethylene glycol) polymer brush for immobilization on glass-type surfaces. Biosens Bioelectron. 2000;15(9–10):473–81.
Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. Molecular conformation in oligo(ethylene glycol)-terminated self-assembled monolayers on gold and silver surfaces determines their ability to resist protein adsorption. J Phys Chem B. 1998;102(2):426–36.
Kunze A, Dilcher M, Abd El Wahed A, Hufert F, Niessner R, Seidel M. On-chip isothermal nucleic acid amplification on flow-based chemiluminescence microarray analysis platform for the detection of viruses and bacteria. Anal Chem. 2015;88(1):898–905.
Wunderlich A, Torggler C, Elsässer D, Lück C, Niessner R, Seidel M. Rapid quantification method for Legionella pneumophila in surface water. Anal Bioanal Chem. 2016;408(9):2203–13.
Sievers S, Cretich M, Gagni P, Ahrens B, Grishina G, Sampson H, et al. Performance of a polymer coated silicon microarray for simultaneous detection of food allergen-specific IgE and IgG4. Clin Exp Allergy. 2017;47(8):1057–68.
Chen TF, Siow KS, Ng PY, Nai MH, Lim CT, Yeop Majlis B. Ageing properties of polyurethane methacrylate and off-stoichiometry thiol-ene polymers after nitrogen and argon plasma treatment. J Appl Polym Sci 2016;133(42).
Seidel M, Dankbar DM, Gauglitz G. A miniaturized heterogeneous fluorescence immunoassay on gold-coated nano-titer plates. Anal Bioanal Chem. 2004;379(7–8):904–12.
Sola L, Damin F, Gagni P, Consonni R, Chiari M. Synthesis of clickable coating polymers by postpolymerization modification: applications in microarray technology. Langmuir. 2016;32(40):10284–95.
Gori A, Cretich M, Vanna R, Sola L, Gagni P, Bruni G, et al. Multiple epitope presentation and surface density control enabled by chemoselective immobilization lead to enhanced performance in IgE-binding fingerprinting on peptide microarrays. Anal Chim Acta. 2017;983:189–97.
Tamarit-López J, Morais S, Puchades R, Maquieira A. Oxygen plasma treated interactive polycarbonate DNA microarraying platform. Bioconjug Chem. 2011;22(12):2573–80.
Li Y, Wang Z, Ou L, Yu HZ. DNA detection on plastic: surface activation protocol to convert polycarbonate substrates to biochip platforms. Anal Chem. 2007;79(2):426–33.
Rowinska M, Kelleher S, Soberon F, Ricco A, Daniels S. Fabrication and characterisation of spin coated oxidised PMMA to provide a robust surface for on-chip assays. J Mater Chem B. 2015;3(1):135–43.
Ahn J, Shin Y-B, Chang W-S, Kim M-G. Sequential patterning of two fluorescent streptavidins assisted by photoactivatable biotin on an aminodextran-coated surface. Colloids Surf B. 2011;87(1):67–72.
Papp K, Holczer E, Kecse-Nagy C, Szittner Z, Lóránd V, Rovero P, et al. Multiplex determination of antigen specific antibodies with cell binding capability in a self-driven microfluidic system. Sensors Actuators B Chem. 2017;238:1092–7.
Jang M, Park CK, Lee NY. Modification of polycarbonate with hydrophilic/hydrophobic coatings for the fabrication of microdevices. Sensors Actuators B Chem. 2014;193:599–607.
Wessig P, Bendig J, Schedler U (2004) Surface-functionalised carrier material, method for the production thereof and solid phase synthesis method. EP1355978B1.
Dankbar DM, Gauglitz G. A study on photolinkers used for biomolecule attachment to polymer surfaces. Anal Bioanal Chem. 2006;386(7–8):1967–74.
Jankowski P, Ogonczyk D, Kosinski A, Lisowski W, Garstecki P. Hydrophobic modification of polycarbonate for reproducible and stable formation of biocompatible microparticles. Lab Chip. 2010;11(4):748–52.
Péter M, Schüler T, Furthner FO, Rensing PA, van Heck GT, Schoo HF, et al. Flexible biochips for detection of biomolecules. Langmuir. 2009;25(9):5384–90.
Schüler T, Kretschmer R, Jessing S, Urban M, Fritzsche W, Möller R, et al. A disposable and cost efficient microfluidic device for the rapid chip-based electrical detection of DNA. Biosens Bioelectron. 2009;25(1):15–21.
Wünscher S, Seise B, Pretzel D, Pollok S, Perelaer J, Weber K, et al. Chip-on-foil devices for DNA analysis based on inkjet-printed silver electrodes. Lab Chip. 2014;14(2):392–401.
Ohlander A, Zilio C, Hammerle T, Zelenin S, Klink G, Chiari M, et al. Genotyping of single nucleotide polymorphisms by melting curve analysis using thin film semi-transparent heaters integrated in a lab-on-foil system. Lab Chip. 2013;13(11):2075–82.
Van Zant P. Microchip fabrication: a practical guide to semiconductor processing. McGraw-Hill; 2001.
Kloth K, Niessner R, Seidel M. Development of an open stand-alone platform for regenerable automated microarrays. Biosens Bioelectron. 2009;24(7):2106–12.
Lengger S, Otto J, Elsässer D, Schneider O, Tiehm A, Fleischer J, et al. Oligonucleotide microarray chip for the quantification of MS2, ΦX174, and adenoviruses on the multiplex analysis platform MCR 3. Anal Bioanal Chem. 2014;406(14):3323–34.
Zhu H, Qian J. Applications of functional protein microarrays in basic and clinical research. Advances in genetics Elsevier; 2012. p. 123–55.
Angenendt P. Progress in protein and antibody microarray technology. Drug Discov Today. 2005;10(7):503–11.
Puetz J, Aegerter M. Dip coating technique. In: Aegerter MA, Mennig M, editors. Sol-Gel Technologies for Glass Producers and Users. Boston: Springer; 2004. p. 37–48.
Sui X, Zapotoczny S, Benetti EM, Schön P, Vancso GJ. Characterization and molecular engineering of surface-grafted polymer brushes across the length scales by atomic force microscopy. J Mater Chem. 2010;20(24):4981–93.
Panzarasa G. The art and science of polymer brushes: recent developments in patterning and characterization approaches. Chimia. 2017;71(6):354–8.
Gong P, Levicky R. DNA surface hybridization regimes. Proc Natl Acad Sci. 2008;105(14):5301–6.
Acknowledgements
The authors thank Dr. Christian Lück (Konsiliarlaboratorium für Legionellen, Dresden University of Technology) for providing the monoclonal antibody 10/6 specific for Legionella pneumophila serogroup 1 strain Bellingham-1 and Prof. Erwin Märtlbauer and Dr. Richard Dietrich (Chair for Hygiene and Technology of Milk, Ludwig-Maximilians-Universität München) for providing the monoclonal antibody mAb 1F7 reactive against clinafloxicin and enrofloxacin. The authors also thank Dr. Thomas Baumann (Institute of Hydrochemistry and Chair of Analytical Chemistry and Water Chemistry, Technical University of Munich) for the advice on image processing and analysis.
Funding
This work was funded by Bundesministerium für Bildung und Forschung (BMBF) – LegioTyper (FKZ: 13N13698) and received partial support from Hanns Seidel Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ABC Highlights: authored by Rising Stars and Top Experts.
Rights and permissions
About this article
Cite this article
Bemetz, J., Kober, C., Meyer, V.K. et al. Succinylated Jeffamine ED-2003 coated polycarbonate chips for low-cost analytical microarrays. Anal Bioanal Chem 411, 1943–1955 (2019). https://doi.org/10.1007/s00216-019-01594-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-01594-8