Abstract
It is relatively easy to collect chromatographic measurements for a large number of analytes, especially with gradient chromatographic methods coupled with mass spectrometry detection. Such data often have a hierarchical or clustered structure. For example, analytes with similar hydrophobicity and dissociation constant tend to be more alike in their retention than a randomly chosen set of analytes. Multilevel models recognize the existence of such data structures by assigning a model for each parameter, with its parameters also estimated from data. In this work, a multilevel model is proposed to describe retention time data obtained from a series of wide linear organic modifier gradients of different gradient duration and different mobile phase pH for a large set of acids and bases. The multilevel model consists of (1) the same deterministic equation describing the relationship between retention time and analyte-specific and instrument-specific parameters, (2) covariance relationships relating various physicochemical properties of the analyte to chromatographically specific parameters through quantitative structure–retention relationship based equations, and (3) stochastic components of intra-analyte and interanalyte variability. The model was implemented in Stan, which provides full Bayesian inference for continuous-variable models through Markov chain Monte Carlo methods.

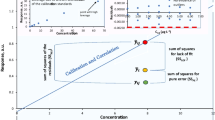
Relationships between log k and MeOH content for acidic, basic, and neutral compounds with different log P. CI credible interval, PSA polar surface area





Similar content being viewed by others
References
Wiczling P, Struck-Lewicka W, Kubik L, Siluk D, Markuszewski MJ, Kaliszan R. The simultaneous determination of hydrophobicity and dissociation constant by liquid chromatography-mass spectrometry. J Pharm Biomed Anal. 2014;94:180–7.
Kubik Ł, Struck-Lewicka W, Kaliszan R, Wiczling P. Simultaneous determination of hydrophobicity and dissociation constant for a large set of compounds by gradient reverse phase high performance liquid chromatography-mass spectrometry technique. J Chromatogr A. 2015;1416:31–7.
Pappa-Louisi A, Nikitas P, Balkatzopoulou P, Malliakas C. Two- and three-parameter equations for representation of retention data in reversed-phase liquid chromatography. J Chromatogr A. 2004;1033(1):29–41.
Rosés M, Subirats X, Bosch E. Retention models for ionizable compounds in reversed-phase liquid chromatography: effect of variation of mobile phase composition and temperature. J Chromatogr A. 2009;1216(10):1756–75.
Andrés A, Téllez A, Rosés M, Bosch E. Chromatographic models to predict the elution of ionizable analytes by organic modifier gradient in reversed phase liquid chromatography. J Chromatogr A. 2012;1247:71–80.
Téllez A, Rosés M, Bosch E. Modeling the retention of neutral compounds in gradient elution RP-HPLC by means of polarity parameter models. Anal Chem. 2009;81(21):9135–45.
Kaliszan R. QSRR: quantitative structure-(chromatographic) retention relationships. Chem Rev. 2007;107(7):3212–46.
Park SH, Haddad PR, Amos RIJ, Talebi M, Szucs R, Pohl CA, et al. Towards a chromatographic similarity index to establish localised quantitative structure-retention relationships for retention prediction. III combination of Tanimoto similarity index, logP, and retention factor ratio to identify optimal analyte training sets for ion chromatography. J Chromatogr A. 2017;1520:107–16.
Tyteca E, Talebi M, Amos R, Park SH, Taraji M, Wen Y, et al. Towards a chromatographic similarity index to establish localized quantitative structure-retention models for retention prediction: Use of retention factor ratio. J Chromatogr A. 2017;1486:50–8.
Daghir-Wojtkowiak E, Wiczling P, Waszczuk-Jankowska M, Kaliszan R, Markuszewski MJ. Multilevel pharmacokinetics-driven modeling of metabolomics data. Metabolomics. 2017;13(3):31.
Wiczling P, Bartkowska-Śniatkowska A, Szerkus O, Siluk D, Rosada-Kurasińska J, Warzybok J, et al. The pharmacokinetics of dexmedetomidine during long-term infusion in critically ill pediatric patients. A Bayesian approach with informative priors. J Pharmacokinet Pharmacodyn. 2016;43(3):315–24.
Lin C, Gelman A, Price PN, Krantz DH. Analysis of local decisions using hierarchical modeling, applied to home radon measurement and remediation. Stat Sci. 1999;14(3):333–7.
Wiczling P, Kaliszan R. How much can we learn from a single chromatographic experiment? A Bayesian perspective. Anal Chem. 2016;88(1):997–1002.
Wiczling P, Kubik Ł, Kaliszan R. Maximum a posteriori Bayesian estimation of chromatographic parameters by limited number of experiments. Anal Chem. 2015;87(14):7241–9.
Wiczling P. Evaluation of sequential Bayesian-based method development procedures for chromatographic problems involving one, two, and three analytes. Sep Sci Plus. 2018;1(2):63–75. https://doi.org/10.1002/sscp.201700037.
Wiczling P, Kawczak P, Nasal A, Kaliszan R. Simultaneous determination of pKa and lipophilicity by gradient RP HPLC. Anal Chem. 2006;78(1):239–49.
Canals I, Portal J, Bosch E, Roses M. Retention of ionizable compounds on HPLC. 4. Mobile-phase pH measurement in methanol/water. Anal Chem. 2000;72(8):1802–9.
Wiczling P, Markuszewski MJ, Kaliszan M, Kaliszan R. pH/organic solvent double-gradient reversed-phase HPLC. Anal Chem. 2005;77(2):449–58.
Snyder LR, Dolan JW. High-performance gradient elution: the practical application of the linear-solvent-strength model. Hoboken: Wiley; 2006.
Nikitas P, Pappa-Louisi A. Expressions of the fundamental equation of gradient elution and a numerical solution of these equations under any gradient profile. Anal Chem. 2005;77(17):5670–7.
Wiczling P, Kaliszan R. Retention time and peak width in the combined pH/organic modifier gradient high performance liquid chromatography. J Chromatogr A. 2010;1217(20):3375–81.
Wiczling P, Kaliszan R. Influence of pH on retention in linear organic modifier gradient RP HPLC. Anal Chem. 2008;80(20):7855–61.
Snyder LR, Kirkland JJ, Dolan JW. Introduction to modern liquid chromatography. 3rd ed. Oxford: Wiley-Blackwell; 2010.
Gelman A. Bayesian data analysis. 2nd ed. Boca Raton: CRC; 2004.
Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B, Betancourt M, et al. Stan: a probabilistic programming language. J Stat Softw. 2017;76(1):1–29.
Gelman A, Hill J. Data analysis using regression and multilevel/hierarchical models. Cambridge: Cambridge University Press; 2007.
McElreath R. Statistical rethinking: a Bayesian course with examples in R and Stan. Boca Raton: CRC; 2016.
Neue U, Phoebe C, Tran K, Cheng Y, Lu Z. Dependence of reversed-phase retention of ionizable analytes on pH, concentration of organic solvent and silanol activity. J Chromatogr A. 2001;925(1-2):49–67.
Hansch C, Leo A, Taft R. A Survey of Hammett substituent constants and resonance and field parameters. Chem Rev. 1991;91(2):165–95.
Gritti F, Guiochon G. Critical contribution of nonlinear chromatography to the understanding of retention mechanism in reversed-phase liquid chromatography. J Chromatogr A. 2005;1099(1-2):1–42.
Gritti F, Guiochon G. Overloaded elution band profiles of ionizable compounds in reversed-phase liquid chromatography: influence of the competition between the neutral and the ionic species. J Sep Sci. 2008;31(21):3657–82.
Haddad PR. Seeking the holy grail—prediction of chromatographic retention based only on chemical structures. LCGC. 2017;35(8):499–502.
Kubik Ł, Wiczling P. Quantitative structure-(chromatographic) retention relationship models for dissociating compounds. J Pharm Biomed Anal. 2016;127:176–83.
Acknowledgement
This project was supported by the National Science Centre, Poland (grant 2015/18/E/ST4/00449).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no competing interests.
Rights and permissions
About this article
Cite this article
Wiczling, P. Analyzing chromatographic data using multilevel modeling. Anal Bioanal Chem 410, 3905–3915 (2018). https://doi.org/10.1007/s00216-018-1061-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1061-3