Abstract
The monitoring of protein biomarkers for the early prediction of cell stress and death is a valuable tool for process characterization and efficient biomanufacturing control. A representative set of six proteins, namely GPDH, PRDX1, LGALS1, CFL1, TAGLN2 and MDH, which were identified in a previous CHO-K1 cell death model using discovery LC-MSE was translated into a targeted liquid chromatography multiple reaction monitoring mass spectrometry (LC-MRM-MS) platform and verified. The universality of the markers was confirmed in a cell growth model for which three Chinese hamster ovary host cell lines (CHO-K1, CHO-S, CHO-DG44) were grown in batch culture in two different types of basal media. LC-MRM-MS was also applied to spent media (n = 39) from four perfusion biomanufacturing series. Stable isotope-labelled peptide analogues and a stable isotope-labelled monoclonal antibody were used for improved protein quantitation and simultaneous monitoring of the workflow reproducibility. Significant increases in protein concentrations were observed for all viability marker proteins upon increased dead cell numbers and allowed for discrimination of spent media with dead cell densities below and above 1 × 106 dead cells/mL which highlights the potential of the selected viability marker proteins in bioprocess control.

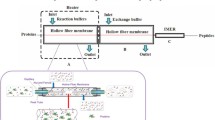
Overview of the LC-MRM-MS workflow for the determination of proteomic markers in conditioned media from the bioreactor that correlate with CHO cell death






Similar content being viewed by others
References
Kumar N, Gammell P, Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture: a summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology. 2007;53(1–3):33–46.
Li F, Vijayasankaran N, Shen AY, Kiss R, Amanullah A. Cell culture processes for monoclonal antibody production. MAbs. 2010;2(5):466–79.
Reinhart D, Damjanovic L, Kaisermayer C, Kunert R. Benchmarking of commercially available CHO cell culture media for antibody production. Appl Microbiol Biotechnol. 2015;99(11):4645–57.
Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22(11):1393–8.
Lee JC, Chang HN, Oh DJ. Recombinant antibody production by perfusion cultures of rCHO cells in a depth filter perfusion system. Biotechnol Prog. 2005;21(1):134–9.
Zydney AL. Perspectives on integrated continuous bioprocessing—opportunities and challenges. Curr Opin Chem Eng. 2015;10:8–13.
Krampe B, Al-Rubeai M. Cell death in mammalian cell culture: molecular mechanisms and cell line engineering strategies. Cytotechnology. 2010;62(3):175–88.
Chan FK, Moriwaki K, De Rosa MJ. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol Biol. 2013;979:65–70.
Crowley LC, Marfell BJ, Christensen ME, Waterhouse NJ. Measuring cell death by trypan blue uptake and light microscopy. Cold Spring Harb Protoc. 2016;2016(7):pdb.prot087155.
Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16(8):1093–107.
Silva MT. Secondary necrosis: the natural outcome of the complete apoptotic program. FEBS Lett. 2010;584(22):4491–9.
Cummings BS, Wills LP, Schnellmann RG. Measurement of cell death in mammalian cells. Current protocols in pharmacology/editorial board, SJ Enna (editor-in-chief) [et al] 0 12. 2004. https://doi.org/10.1002/0471141755.ph0471141208s0471141725.
Braasch K, Nikolic-Jaric M, Cabel T, Salimi E, Bridges GE, Thomson DJ, et al. The changing dielectric properties of CHO cells can be used to determine early apoptotic events in a bioprocess. Biotechnol Bioeng. 2013;110(11):2902–14.
Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, Curi R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem Funct. 2003;21(1):1–9.
Schmidberger T, Gutmann R, Bayer K, Kronthaler J, Huber R. Advanced online monitoring of cell culture off-gas using proton transfer reaction mass spectrometry. Biotechnol Prog. 2014;30(2):496–504.
Zagari F, Jordan M, Stettler M, Broly H, Wurm FM. Lactate metabolism shift in CHO cell culture: the role of mitochondrial oxidative activity. New Biotechnol. 2013;30(2):238–45.
Albrecht S, Kaisermayer C, Gallagher C, Farrell A, Lindeberg A, Bones J. Proteomics in biomanufacturing control: protein dynamics of CHO-K1 cells and conditioned media during apoptosis and necrosis. Biotechnol Bioeng. 2018;1–12. https://doi.org/10.1002/bit.26563.
Doneanu CE, Xenopoulos A, Fadgen K, Murphy J, Skilton SJ, Prentice H, et al. Analysis of host-cell proteins in biotherapeutic proteins by comprehensive online two-dimensional liquid chromatography/mass spectrometry. MAbs. 2012;4(1):24–44.
Wang X, Schomogy T, Wells K, Mozier N. Improved HCP quantitation by minimizing antibody cross-reactivity to target proteins. BioProcess Int. 2010:18–24.
Liebler DC, Zimmerman LJ. Targeted quantitation of proteins by mass spectrometry. Biochemistry. 2013;52(22):3797–806.
Muqaku B, Slany A, Bileck A, Kreutz D, Gerner C. Quantification of cytokines secreted by primary human cells using multiple reaction monitoring: evaluation of analytical parameters. Anal Bioanal Chem. 2015;407(21):6525–36.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8.
Woolley JF, Al-Rubeai M. The isolation and identification of a secreted biomarker associated with cell stress in serum-free CHO cell culture. Biotechnol Bioeng. 2009;104(3):590–600.
Mei W, Peng Z, Lu M, Liu C, Deng Z, Xiao Y, et al. Peroxiredoxin 1 inhibits the oxidative stress induced apoptosis in renal tubulointerstitial fibrosis. Nephrology (Carlton). 2015;20(11):832–42.
Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol. 2003;5(12):1083–9.
Teasdale A, Jahn M, Bailey S, Feilden A, Taylor G, Corcoran ML, et al. Controlled extraction studies applied to polyvinyl chloride and polyethylene materials: conclusions from the ELSIE controlled extraction pilot study. AAPS PharmSciTech. 2015;16(3):664–74.
Lee SM, Kim JH, Cho EJ, Youn HD. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell Death Differ. 2009;16(5):738–48.
Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31(8):759–65.
Wei YY, Naderi S, Meshram M, Budman H, Scharer JM, Ingalls BP, et al. Proteomics analysis of chinese hamster ovary cells undergoing apoptosis during prolonged cultivation. Cytotechnology. 2011;63(6):663–77.
Zhang H, Liu Q, Zimmerman LJ, Ham AJ, Slebos RJ, Rahman J, et al. Methods for peptide and protein quantitation by liquid chromatography-multiple reaction monitoring mass spectrometry. Mol Cell Proteomics. 2011;10(6):M110.006593.
Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–41.
Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581(19):3652–7.
Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15(5):767–76.
Acknowledgements
The authors acknowledge support from Enterprise Ireland under the Innovation Partnership funding program, with grant reference IP/2014/0309, co-funded by the European Union through the European Regional Development Fund (ERDF) 2014-2020 program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Albrecht, S., Kaisermayer, C., Reinhart, D. et al. Multiple reaction monitoring targeted LC-MS analysis of potential cell death marker proteins for increased bioprocess control. Anal Bioanal Chem 410, 3197–3207 (2018). https://doi.org/10.1007/s00216-018-1029-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1029-3