Abstract
The highly multiplexed polymerase chain reaction (PCR) assays used for forensic human identification perform best when used with an accurately determined quantity of input DNA. To help ensure the reliable performance of these assays, we are developing a certified reference material (CRM) for calibrating human genomic DNA working standards. To enable sharing information over time and place, CRMs must provide accurate and stable values that are metrologically traceable to a common reference. We have shown that droplet digital PCR (ddPCR) limiting dilution end-point measurements of the concentration of DNA copies per volume of sample can be traceably linked to the International System of Units (SI). Unlike values assigned using conventional relationships between ultraviolet absorbance and DNA mass concentration, entity-based ddPCR measurements are expected to be stable over time. However, the forensic community expects DNA quantity to be stated in terms of mass concentration rather than entity concentration. The transformation can be accomplished given SI-traceable values and uncertainties for the number of nucleotide bases per human haploid genome equivalent (HHGE) and the average molar mass of a nucleotide monomer in the DNA polymer. This report presents the considerations required to establish the metrological traceability of ddPCR-based mass concentration estimates of human nuclear DNA.

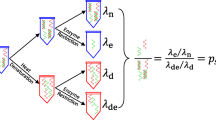
The roots of metrological traceability for human nuclear DNA mass concentration results. Values for the factors in blue must be established experimentally. Values for the factors in red have been established from authoritative source materials. HHGE stands for “haploid human genome equivalent”; there are two HHGE per diploid human genome.

Similar content being viewed by others
References
Kline MC, Duewer DL, Redman JW, Butler JM. NIST Mixed Stain Study #3: DNA quantitation accuracy and its influence on short tandem repeat multiplex signal intensity. Anal Chem. 2003;75(10):2463–9.
Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13(3):444–9.
Kline MC, Duewer DL. Evaluating Droplet Digital Polymerase Chain Reaction for the Quantification of Human Genomic DNA: Lifting the Traceability Fog. Anal Chem. 2017;89(8):4648–54. https://doi.org/10.1021/acs.analchem.7b00240.
Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual (Fourth Edition). Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2012.
De Bièvre P, Dybkaer R, Fajgelj A, Hibbert DB. Metrological traceability of measurement results in chemistry: Concepts and implementation. Pure Appl Chem. 2011;83(10):1873-935. https://doi.org/10.1351/PAC-REP-07-09-39.
Griffiths KR, Burke DG, Emslie KR. Quantitative polymerase chain reaction: a framework for improving the quality of results and estimating uncertainty of measurement. Anal. Methods. 2011;3:2201–11. https://doi.org/10.1039/c1ay05069a.
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal Chem. 2012;84:1003–11. https://doi.org/10.1021/ac202578x.
Jacobs BKM, Goetghebeur E, Clement L. Impact of variance components on reliability of absolute quantification using digital PCR. BMC Bioinf. 2014;15:283. https://doi.org/10.1186/1471-2105-15-283.
Deprez L, Corbisier P, Kortekaas A-M, Mazoua S, Hidalgo RB, Trapmann S, et al. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol Detect Quantif. 2016;9:29–39. https://doi.org/10.1016/j.bdq.2016.08.002.
Corbisier P, Pinheiro L, Mazoua S, Kortekaas A-M, Chung PYJ, Gerganova T, et al. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal Bioanal Chem. 2015;407(7):1831–40. https://doi.org/10.1007/s00216-015-8458-z.
Dong LH, Meng Y, Sui ZW, Wang J, Wu LQ, Fu BQ. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci Rep. 2015;5:13174. https://doi.org/10.1038/srep13174.
Dagata JA, Farkas N, Kramer JA. Method for measuring the volume of nominally 100 μm diameter spherical water-in-oil emulsion droplets. NIST Special Publication 260–184. 2016. https://doi.org/10.6028/NIST.SP.260-184
Košir AB, Divieto C, Pavšič J, Pavarelli S, Dobnik D, Dreo T, et al. Droplet volume variability as a critical factor for accuracy of absolute quantification using droplet digital PCR. Anal Bioanal Chem. 2017; https://doi.org/10.1007/s00216-017-0625-y.
JCGM 100:2008. Evaluation of measurement data — Guide to the expression of uncertainty in measurement (GUM). Joint Committee for Guides in Metrology. Sevres, France. 2008. See Section 4.3.7. https://www.bipm.org/en/publications/guides/#gum
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. https://doi.org/10.1093/nar/16.3.1215.
CDC/NIH: Biosafety in Microbiological and Biomedical Laboratories, 5th ed.; HHS publication No. (CDC) 21–1112; Chosewood, L.C.; Wilson, D.E.; Eds.; US Government Printing Office: Washington, DC (2009) http://www.cdc.gov/biosafety/publications/bmbl5/index.htm
NCBI. Standard Nucleotide BLAST. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
Kline MC, Romsos EL, Duewer DL. Evaluating Digital PCR for the Quantification of Human Genomic DNA: Accessible Amplifiable Targets. Anal Chem. 2016;88(4):2132–9. https://doi.org/10.1021/acs.analchem.5b03692.
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. https://doi.org/10.1038/nature15393.
Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–64. https://doi.org/10.1101/gr.213611.116.
Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature. 2012;13:36–46. https://doi.org/10.1038/nrg3117.
Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–11. https://doi.org/10.1038/nature13907.
Memon AA, Zöller B, Hedelius A, Wang X, Stenman E, Sundquist J, et al. Quantification of mitochondrial DNA copy number in suspected cancer patients by a well optimized ddPCR method. Biomol Detect Quantif. 2017;13:32–9. https://doi.org/10.1016/j.bdq.2017.08.001.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. https://doi.org/10.1038/13779.
Thomas RA, Krishan A, Robinson DM, Sams C, Costa F. NASA/American Cancer Society High-Resolution Flow Cytometry Project—I. Cytometry. 2001;43:2–11. https://doi.org/10.1002/1097-0320(20010101)43:1<2::AID-CYTO1012>3.0.CO;2-J.
Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51A:127–8. https://doi.org/10.1002/cyto.a.10013.
Meija J, Coplen TB, Berglund M, Brand WA, De Bièvre P, Gröning M, et al. Atomic weights of the elements 2013. Pure Appl Chem. 2016;88:265–91. https://doi.org/10.1515/pac-2015-0305.
Possolo A, van der Veen AMH, Meija J, Hibbert DB. Interpreting and propagating the uncertainty of the standard atomic weights (IUPAC Technical Report). Pure Appl Chem 2017; aop. https://doi.org/10.1515/pac-2016-0402.
Vinogradov AE. Measurement by flow cytometry of genomic AT/GC ratio and genome size. Cytometry. 1994;16:34–40. https://doi.org/10.1002/cyto.990160106.
Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33. https://doi.org/10.1038/nature17640.
Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709-21.
Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, et al. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLoS One. 2012;7(7):e41361. https://doi.org/10.1371/journal.pone.0041361.
Acknowledgments
We thank our NIST colleague Justin Zook for interpreting the information provided by The Genome Reference Consortium and an anonymous Referee for insightful suggestions and corrections. This work was supported in part by the NIST Special Programs Office project Forensic DNA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animal rights
All work presented has been reviewed and approved by the National Institute of Standards and Technology Human Subjects Protections Office. This study was determined to be “not human subjects research” (often referred to as research not involving human subjects) as defined in U. S. Department of Commerce Regulations, 15 CFR 27, also known as the Common Rule (45 CFR 46, Subpart A), for the Protection of Human Subjects by the NIST Human Subjects Protection Office and therefore not subject to oversight by the NIST Institutional Review Board.
Conflict of interest
The authors declare that they have no conflict of interest.
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this report to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Electronic supplementary material
ESM 1
(PDF 841 kb)
Rights and permissions
About this article
Cite this article
Duewer, D.L., Kline, M.C., Romsos, E.L. et al. Evaluating droplet digital PCR for the quantification of human genomic DNA: converting copies per nanoliter to nanograms nuclear DNA per microliter. Anal Bioanal Chem 410, 2879–2887 (2018). https://doi.org/10.1007/s00216-018-0982-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-0982-1