Abstract
The 3′-polyadenosine (poly A) tail of in vitro transcribed (IVT) mRNA was studied using liquid chromatography coupled to mass spectrometry (LC-MS). Poly A tails were cleaved from the mRNA using ribonuclease T1 followed by isolation with dT magnetic beads. Extracted tails were then analyzed by LC-MS which provided tail length information at single-nucleotide resolution. A 2100-nt mRNA with plasmid-encoded poly A tail lengths of either 27, 64, 100, or 117 nucleotides was used for these studies as enzymatically added poly A tails showed significant length heterogeneity. The number of As observed in the tails closely matched Sanger sequencing results of the DNA template, and even minor plasmid populations with sequence variations were detected. When the plasmid sequence contained a discreet number of poly As in the tail, analysis revealed a distribution that included tails longer than the encoded tail lengths. These observations were consistent with transcriptional slippage of T7 RNAP taking place within a poly A sequence. The type of RNAP did not alter the observed tail distribution, and comparison of T3, T7, and SP6 showed all three RNAPs produced equivalent tail length distributions. The addition of a sequence at the 3′ end of the poly A tail did, however, produce narrower tail length distributions which supports a previously described model of slippage where the 3′ end can be locked in place by having a G or C after the poly nucleotide region.
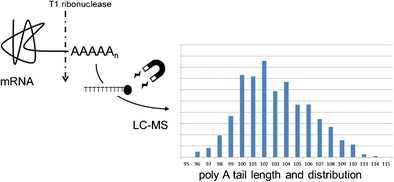
Determination of mRNA poly A tail length using magnetic beads and LC-MS.








Similar content being viewed by others
References
Harrison PF, Powell DR, Clancy JL, Preiss T, Boag PR, Traven A, et al. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA. 2015;21(8):1502–10.
Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19(6):568–76.
Moqtaderi Z, Geisberg JV, Struhl K. Secondary structures involving the poly(A) tail and other 3′ sequences are major determinants of mRNA isoform stability in yeast. Microb Cell. 2014;1(4):137–9.
Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508(7494):66–71.
Ratcliffe CD, Sahgal P, Parachoniak CA, Ivaska J, Park M. Regulation of cell migration and beta1 integrin trafficking by the endosomal adaptor GGA3. Traffic. 2016;17(6):670–88.
Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13(7):982–97.
Peng J, Murray EL, Schoenberg DR. In vivo and in vitro analysis of poly(A) length effects on mRNA translation. Methods Mol Biol. 2008;419:215–30.
Koski GK, Kariko K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J Immunol. 2004;172(7):3989–93.
Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9(2):659–70.
Bernstein P, Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989;14(9):373–7.
Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell. 2014;53(6):1044–52.
Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3' RACE applications. RNA. 2012;18(6):1289–95.
Murray EL, Schoenberg DR. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 2008;448:483–504.
Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35(19):e132.
Salles FJ, Strickland S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 1995;4(6):317–21.
Minasaki R, Rudel D, Eckmann CR. Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA Biol. 2014;11(2):111–23.
Muddiman DC, Null AP, Hannis JC. Precise mass measurement of a double-stranded 500 base-pair (309 kDa) polymerase chain reaction product by negative ion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 1999;13(12):1201–4.
Hannis JC, Muddiman DC. Detection of double-stranded PCR amplicons at the attomole level electrosprayed from low nanomolar solutions using FT-ICR mass spectrometry. Fresenius J Anal Chem. 2001;369(3–4):246–51.
Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, et al. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol. 2008;6(7):553–8.
Wolk DM, Kaleta EJ, Wysocki VH. PCR-electrospray ionization mass spectrometry: the potential to change infectious disease diagnostics in clinical and public health laboratories. J Mol Diagn. 2012;14(4):295–304.
Beverly M, Dell A, Parmar P, Houghton L. Label-free analysis of mRNA capping efficiency using RNase H probes and LC-MS. Anal Bioanal Chem. 2016;408(18):5021–30.
Gilar M. Analysis and purification of synthetic oligonucleotides by reversed-phase high-performance liquid chromatography with photodiode array and mass spectrometry detection. Anal Biochem. 2001;298(2):196–206.
Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal Chem. 1997;69(7):1320–5.
Huber CG, Oberacher H. Analysis of nucleic acids by on-line liquid chromatography-mass spectrometry. Mass Spectrom Rev. 2001;20(5):310–43.
Hail ME, Elliott B, Anderson K. High-throughput analysis of oligonucleotides using automated electrospray ionization mass spectrometry. Am Biotechnol Lab. 2004;22:12–3.
Haukanes BI, Kvam C. Application of magnetic beads in bioassays. Biotechnology (N Y). 1993;11(1):60–3.
Berensmeier S. Magnetic particles for the separation and purification of nucleic acids. Appl Microbiol Biotechnol. 2006;73(3):495–504.
Adams NM, Bordelon H, Wang KK, Albert LE, Wright DW, Haselton FR. Comparison of three magnetic bead surface functionalities for RNA extraction and detection. ACS Appl Mater Interfaces. 2015;7(11):6062–9.
Blower MD, Jambhekar A, Schwarz DS, Toombs JA. Combining different mRNA capture methods to analyze the transcriptome: analysis of the Xenopus laevis transcriptome. PLoS One. 2013;8(10):e77700.
Cabada MO, Darnbrough C, Ford PJ, Turner PC. Differential accumulation of two size classes of poly(A) associated with messenger RNA during oogenesis in Xenopus laevis. Dev Biol. 1977;57(2):427–39.
Park JE, Yi H, Kim Y, Chang H, Kim VN. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell. 2016;62(3):462–71.
Jacobsen N, Nielsen PS, Jeffares DC, Eriksen J, Ohlsson H, Arctander P, et al. Direct isolation of poly(A)+ RNA from 4 M guanidine thiocyanate-lysed cell extracts using locked nucleic acid-oligo(T) capture. Nucleic Acids Res. 2004;32(7):e64.
Phelan D, Hondorp K, Choob M, Efimov V, Fernandez J. Messenger RNA isolation using novel PNA analogues. Nucleosides Nucleotides Nucleic Acids. 2001;20(4–7):1107–11.
Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–17.
Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem. 1995;270(11):6298–307.
Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18(12):3529–35.
Molodtsov V, Anikin M, McAllister WT. The presence of an RNA:DNA hybrid that is prone to slippage promotes termination by T7 RNA polymerase. J Mol Biol. 2014;426(18):3095–107.
Macdonald LE, Zhou Y, McAllister WT. Termination and slippage by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;232(4):1030–47.
Anikin M, Molodtsov V, Temiakov D, McAllister WT. Transcript slippage and recoding. In: Atkins JF, Gesteland RF, editors. Recoding: expansion of decoding rules enrishes gene expression, Nucleic acid and molecular biology: Springer; 2010. p. 409–32.
Elango N, Elango S, Shivshankar P, Katz MS. Optimized transfection of mRNA transcribed from a d(a/T)100 tail-containing vector. Biochem Biophys Res Commun. 2005;330(3):958–66.
Grier AE, Burleigh S, Sahni J, Clough CA, Cardot V, Choe DC, et al. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol Ther Nucleic Acids. 2016;5:e306.
Mockey M, Goncalves C, Dupuy FP, Lemoine FM, Pichon C, Midoux P. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with poly(A) chains in cis and in trans for a high protein expression level. Biochem Biophys Res Commun. 2006;340(4):1062–8.
Funding
Support for this work was provided by DARPA grant HR-0011-13-3-0003.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Disclaimer
The views, opinions, and/or finding expressed are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the US Government.
Electronic supplementary material
ESM 1
(PDF 1.04 mb)
Rights and permissions
About this article
Cite this article
Beverly, M., Hagen, C. & Slack, O. Poly A tail length analysis of in vitro transcribed mRNA by LC-MS. Anal Bioanal Chem 410, 1667–1677 (2018). https://doi.org/10.1007/s00216-017-0840-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0840-6




