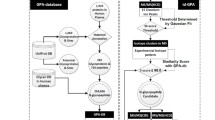
Abstract
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide, largely because of difficulties in early diagnosis. Despite accumulating evidence indicating that aberrant glycosylation is associated with GC, site-specific localization of the glycosylation to increase specificity and sensitivity for clinical use is still an analytical challenge. Here, we created an analytical platform with a targeted glycoproteomic approach for GC biomarker discovery. Unlike the conventional glycomic approach with untargeted mass spectrometric profiling of released glycan, our platform is characterized by three key features: it is a target-protein-specific, glycosylation-site-specific, and structure-specific platform with a one-shot enzyme reaction. Serum haptoglobin enriched by immunoaffinity chromatography was subjected to multispecific proteolysis to generate site-specific glycopeptides and to investigate the macroheterogeneity and microheterogeneity. Glycopeptides were identified and quantified by nano liquid chromatography–mass spectrometry and nano liquid chromatography–tandem mass spectrometry. Ninety-six glycopeptides, each corresponding to a unique glycan/glycosite pairing, were tracked across all cancer and control samples. Differences in abundance between the two groups were marked by particularly high magnitudes. Three glycopeptides exhibited exceptionally high control-to-cancer fold changes along with receiver operating characteristic curve areas of 1.0, indicating perfect discrimination between the two groups. From the results taken together, our platform, which provides biological information as well as high sensitivity and reproducibility, may be useful for GC biomarker discovery.

Graphical abstract
ᅟ






Similar content being viewed by others
References
Hart GW, Copeland RJ. Glycomics hits the big time. Cell. 2010;143(5):672–6.
Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Silva DV, Fuchser J, et al. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J Mass Spectrom. 2013;48(6):627–39.
Ruhaak LR, H-W U, Beekman M, Hokke CH, Westendorp RG, Houwing-Duistermaat J, et al. Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J Proteome Res. 2011;10(4):1667–74.
Hua S, Saunders M, Dimapasoc LM, Jeong SH, Kim BJ, Kim S, et al. Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J Proteome Res. 2014;13(2):961–8.
Bondt A, Selman MH, Deelder AM, Hazes JM, Willemsen SP, Wuhrer M, et al. Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J Proteome Res. 2013;12(10):4522–31.
Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–55.
Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–71.
Hua S, Williams CC, Dimapasoc LM, Ro GS, Ozcan S, Miyamoto S, et al. Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. J Chromatogr A. 2013;1279:58–67. https://doi.org/10.1016/j.chroma.2012.12.079
Ozcan S, Barkauskas DA, Ruhaak LR, Torres J, Cooke CL, An HJ, et al. Serum glycan signatures of gastric cancer. Cancer Prev Res. 2014;7(2):226–35.
Lee SH, Jeong S, Lee J, Yeo IS, MJ O, Kim U, et al. Glycomic profiling of targeted serum haptoglobin for gastric cancer using nano LC/MS and LC/MS/MS. Mol Biosyst. 2016;12(12):3611–21.
Campos D, Freitas D, Gomes J, Magalhães A, Steentoft C, Gomes C, et al. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol Cell Proteomics. 2015;14(6):1616–29.
Shah P, Wang X, Yang W, Eshghi ST, Sun S, Hoti N, et al. Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Mol Cell Proteomics. 2015;14(10):2753–63.
Hua S, An HJ. Glycoscience aids in biomarker discovery. BMB Rep. 2012;45(6):323–30.
Matsuda A, Kuno A, Matsuzaki H, Kawamoto T, Shikanai T, Nakanuma Y, et al. Glycoproteomics-based cancer marker discovery adopting dual enrichment with Wisteria floribunda agglutinin for high specific glyco-diagnosis of cholangiocarcinoma. J Proteomics. 2013;85:1–11.
Kaji H, Ocho M, Togayachi A, Kuno A, Sogabe M, Ohkura T, et al. Glycoproteomic discovery of serological biomarker candidates for HCV/HBV infection-associated liver fibrosis and hepatocellular carcinoma. J Proteome Res. 2013;12(6):2630–40.
Shang S, Li W, Qin X, Zhang S, Liu Y. Aided diagnosis of hepatocellular carcinoma using serum fucosylated haptoglobin ratios. J Cancer. 2017;8(5):887.
Sun S, Wang Q, Zhao F, Chen W, Li Z. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One. 2011;6(7):e22844.
Balla-Jhagjhoorsingh SS, Corti D, Heyndrickx L, Willems E, Vereecken K, Davis D, et al. The N276 glycosylation site is required for HIV-1 neutralization by the CD4 binding site specific HJ16 monoclonal antibody. PLoS One. 2013;8(7):e68863.
Thaysen-Andersen M, Packer NH. Advances in LC–MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N-and O-glycoproteome. Biochim Biophys Acta. 2014;1844(9):1437–52.
Dallas DC, Martin WF, Hua S, German JB. Automated glycopeptide analysis—review of current state and future directions. Brief Bioinform. 2012;14(3):361–74.
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomark Prev. 2016;25(1):16–27.
Miki K, Morita M, Sasajima M, Hoshina R, Kanda E, Urita Y. Usefulness of gastric cancer screening using the serum pepsinogen test method. Am J Gastroenterol. 2003;98(4):735–9.
Shimoyama T, Aoki M, Sasaki Y, Matsuzaka M, Nakaji S, Fukuda SABC. screening for gastric cancer is not applicable in a Japanese population with high prevalence of atrophic gastritis. Gastric Cancer. 2012;15(3):331–4.
Chung SJ, Park MJ, Kang SJ, Kang HY, Chung GE, Kim SG, et al. Effect of annual endoscopic screening on clinicopathologic characteristics and treatment modality of gastric cancer in a high-incidence region of Korea. Int J Cancer. 2012;131(10):2376–84.
Kim BJ, Heo C, Kim BK, Kim JY, Kim JG. Effectiveness of gastric cancer screening programs in South Korea: organized vs opportunistic models. World J Gastroenterol. 2013;19(5):736.
Langlois MR, Delanghe JR. Biological and clinical significance of haptoglobin polymorphism in humans. Clin Chem. 1996;42(10):1589–600.
Sadrzadeh SM, Saffari Y, Bozorgmehr J. Haptoglobin phenotypes in epilepsy. Clin Chem. 2004;50(6):1095–7. https://doi.org/10.1373/clinchem.2003.028001
Zhang S, Shang S, Li W, Qin X, Liu Y. Insights on N-glycosylation of human haptoglobin and its association with cancers. Glycobiology. 2016;26(7):684–92.
Kim J-H, Lee SH, Choi S, Kim U, Yeo IS, Kim SH, et al. Direct analysis of aberrant glycosylation on haptoglobin in patients with gastric cancer. Oncotarget. 2017;8(7):11094.
Hua S, Hu CY, Kim BJ, Totten SM, Oh MJ, Yun N, et al. Glyco-analytical multispecific proteolysis (Glyco-AMP): a simple method for detailed and quantitative glycoproteomic characterization. J Proteome Res. 2013;12(10):4414-4423. https://doi.org/10.1021/pr400442y.
ZL W, Ethen C, Hickey GE, Jiang W. Active 1918 pandemic flu viral neuraminidase has distinct N-glycan profile and is resistant to trypsin digestion. Biochem Biophys Res Commun. 2009;379(3):749–53.
An HJ, Tillinghast JS, Woodruff DL, Rocke DM, Lebrilla CB. A new computer program (GlycoX) to determine simultaneously the glycosylation sites and oligosaccharide heterogeneity of glycoproteins. J Proteome Res. 2006;5(10):2800–8.
Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, et al. Automated assignments of N-and O-site specific glycosylation with extensive glycan heterogeneity of glycoprotein mixtures. Anal Chem. 2013;85(12):5666–75.
Kronewitter SR, An HJ, De Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9(11):2986–94.
Hua S, MJ O, Ozcan S, Seo YS, Grimm R, An HJ. Technologies for glycomic characterization of biopharmaceutical erythropoietins. Trends Anal Chem. 2015;68:18–27.
Stavenhagen K, Kolarich D, Wuhrer M. Clinical glycomics employing graphitized carbon liquid chromatography–mass spectrometry. Chromatographia. 2015;78(5-6):307–20.
An HJ, Peavy TR, Hedrick JL, Lebrilla CB. Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Anal Chem. 2003;75(20):5628–37.
Juhasz P, Martin SA. The utility of nonspecific proteases in the characterization of glycoproteins by high-resolution time-of-flight mass spectrometry. Int J Mass Spectrom Ion Processes. 1997;169:217–30.
Cancilla MT, Wong AW, Voss LR, Lebrilla CB. Fragmentation reactions in the mass spectrometry analysis of neutral oligosaccharides. Anal Chem. 1999;71(15):3206–18.
Buck CA, Glick MC, Warren L. Glycopeptides from the surface of control and virus-transformed cells. Science. 1971;172(3979):169–71.
Bresalier RS, Byrd JC, Tessler D, Lebel J, Koomen J, Hawke D, et al. A circulating ligand for galectin-3 is a haptoglobin-related glycoprotein elevated in individuals with colon cancer. Gastroenterology. 2004;127(3):741–8.
Sharpe-Timms KL, Zimmer RL, Ricke EA, Piva M, Horowitz GM. Endometriotic haptoglobin binds to peritoneal macrophages and alters their function in women with endometriosis. Fertil Steril. 2002;78(4):810–9.
Park S-Y, Lee S-H, Kawasaki N, Itoh S, Kang K, Ryu SH, et al. α1-3/4 fucosylation at Asn 241 of β-haptoglobin is a novel marker for colon cancer: a combinatorial approach for development of glycan biomarkers. Int J Cancer. 2012;130(10):2366–76.
Park S-Y, Yoon S-J, Jeong Y-T, Kim J-M, Kim J-Y, Bernert B, et al. N-glycosylation status of β-haptoglobin in sera of patients with colon cancer, chronic inflammatory diseases and normal subjects. Int J Cancer. 2010;126(1):142–55. https://doi.org/10.1002/ijc.24685
Carlsson MC, Cederfur C, Schaar V, Balog CI, Lepur A, Touret F, et al. Galectin-1-binding glycoforms of haptoglobin with altered intracellular trafficking, and increase in metastatic breast cancer patients. PLoS One. 2011;6(10):e26560.
Acknowledgements
This work was supported by Korea Basic Science Institute grant (T37413) and by the Institute for Basic Science (IBS-R001-D1-2017-a00).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participat
Human sera were collected by the National Biobank of Korea by means of a standardized protocol approved by the Ethics Committees of Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea (KH2010-15). Informed consent was provided by all individuals involved in the study.
Electronic supplementary material
ESM 1
(PDF 850 kb)
Rights and permissions
About this article
Cite this article
Lee, J., Hua, S., Lee, S.H. et al. Designation of fingerprint glycopeptides for targeted glycoproteomic analysis of serum haptoglobin: insights into gastric cancer biomarker discovery. Anal Bioanal Chem 410, 1617–1629 (2018). https://doi.org/10.1007/s00216-017-0811-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0811-y