Abstract
A versatile and universal DNA sensing platform is presented based on enzyme-DNA binding protein tags conjugates and simple DNA nanostructures. Two enzyme conjugates were thus prepared, with horseradish peroxidase linked to the dimeric single-chain bacteriophage Cro repressor protein (HRP-scCro) and glucose oxidase linked to the dimeric headpiece domain of Escherichia coli LacI repressor protein (GOx-dHP), and used in conjunction with a hybrid ssDNA-dsDNA detection probe. This probe served as a simple DNA nanostructure allowing first for target recognition through its target-complementary single-stranded DNA (ssDNA) part and then for signal generation after conjugate binding on the double-stranded DNA (dsDNA) containing the specific binding sites for the dHP and scCro DNA binding proteins. The DNA binding proteins chosen in this work have different sequence specificity, high affinity, and lack of cross-reactivity. The proposed sensing system was validated for the detection of model target ssDNA from high-risk human papillomavirus (HPV16) and the limits of detection of 45, 26, and 21 pM were achieved using the probes with scCro/dHP DNA binding sites ratio of 1:1, 2:1, and 1:2, respectively. The performance of the platform in terms of limit of detection was comparable to direct HRP systems using target-specific oligonucleotide-HRP conjugates. The ratio of the two enzymes can be easily manipulated by changing the number of binding sites on the detection probe, offering further optimization possibilities of the signal generation step. Moreover, since the signal is obtained in the absence of externally added hydrogen peroxide, the described platform is compatible with paper-based assays for molecular diagnostics applications. Finally, just by changing the ssDNA part of the detection probe, this versatile nucleic acid platform can be used for the detection of different ssDNA target sequences or in a multiplex detection configuration without the need to change any of the conjugates.

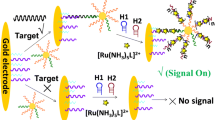
DNA sensing platform based on an immobilized ssDNA capture probe and a hybrid ssDNA-dsDNA detection probe that specifically hybridize with the ssDNA target. The hybrid ssDNA-dsDNA detection probe also provides the binding sites for the enzyme-DNA binding protein conjugates (HRP-scCro and GOx-dHP) that generate the colorimetric signal





Similar content being viewed by others
References
Jain KK. Nanodiagnostics: application of nanotechnology in molecular diagnostics. Expert Rev Mol Diagn. 2014;3:153–61.
Shields D, Dhabale S. 2014. https://www.alliedmarketresearch.com/DNA-diagnostics-market. Accessed 10 Oct 2016.
Kiechle FL. Molecular pathology and infectious diseases. In: Grody WW, Nakaruma RM, Strom CM, Kiechle FL, editors. Molecular diagnostics: techniques and applications for the clinical laboratory. London: Academic Press; 2010. p. 99–106.
Pieretti M. Signal amplification methods in molecular diagnostics. In: Grody WW, Nakaruma RM, Strom CM, Kiechle FL, editors. Molecular diagnostics: techniques and applications for the clinical laboratory. London: Academic Press; 2010. p. 15–9.
Aktas GB, Skouridou V, Masip L. Novel signal amplification approach for HRP-based colorimetric genosensors using DNA binding protein tags. Biosens Bioelectron. 2015;74:1005–10.
Ariga K, Ji Q, Mori T, Naito M, Yamauchi Y, Abe H, Hill JP. Enzyme nanoarchitectonics: organization and device application. Chem Soc Rev. 2013;42:6322–45.
Riccardi CM, Mistri D, Hart O, Anuganti M, Lin Y, Kasi RM, Kumar CV. Covalent interlocking of glucose oxidase and peroxidase in the voids of paper: enzyme–polymer “spider webs”. Chem Commun. 2016;52:2593–6.
Küchler A, Yoshimoto M, Luginbühl S, Mavelli F, Walde P. Enzymatic reactions in confined environments. Nat Nanotechnol. 2016;11:409–20.
Wu D, Ren X, Hu L, Fan D, Zheng Y, Wei Q. Electrochemical aptasensor for the detection of adenosine by using PdCu@MWCNTs-supported bienzymes as labels. Biosens Bioelectron. 2015;74:391–7.
Lai YH, Lee CC, King CC, Chuang MC, Ho JAA. Exploitation of stem-loop DNA as a dual-input gene sensing platform: extension to subtyping of influenza A viruses. Chem Sci. 2014;5:4082–90.
Xin L, Zhou C, Yang Z, Liu D. Regulation of an enzyme cascade reaction by a DNA machine. Small. 2013;9:3088–91.
Civit L, Fragoso A, Hölters S, Dürst M, O’Sullivan CK. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal Chim Acta. 2012;715:93–8.
Gao F, Courjean O, Mano N. An improved glucose/O2 membrane-less biofuel cell through glucose oxidase purification. Biosens Bioelectron. 2009;25:356–61.
Hudson JM, Fried MG. Co-operative interactions between the catabolite gene activator protein and the lac repressor at the lactose promoter. J Mol Biol. 1990;214:381–96.
Kalodimos CG, Folkers GE, Boelens R, Kaptein R. Strong DNA binding by covalently linked dimeric Lac headpiece: evidence for the crucial role of the hinge helices. Proc Natl Acad Sci U S A. 2001;98:6039–44.
Spronk CA, Folkers GE, Noordman AM, Wechselberger R, van den Brink N, Boelens, Kaptein R. Hinge-helix formation and DNA bending in various lac repressor-operator complexes. EMBO J. 1999;18(22):6472–80.
Kohen A, Jonsson T, Klinman JP. Effect of protein glycosylation on catalysis: changes in hydrogen tunneling and enthalpy of activation in the glucose oxidase reaction. Biochemistry. 1997;36:2603–11.
Munteanu MG, Vlahovicek K, Parthasaraty S, Simon I, Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem Sci. 1998;23:341–6.
Manera M, Miro M, Estela JM, Cerdá V. A multisyringe flow injection system with immobilized glucose oxidase based on homogeneous chemiluminescence detection. Anal Chim Acta. 2004;508:23–30.
Wilner OI, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat Nanotechnol. 2009;4:249–54.
Freeman R, Sharon E, Teller C, Wilner I. Control of biocatalytic transformations by programmed DNA assemblies. Chem Eur J. 2010;16:3690–8.
Frías IAM, Avelino KYPS, Silva RR, Andrade CAS, Oliveira MDL. Trends in biosensors for HPV: identification and diagnosis. J Sens. 2015; doi:10.1155/2015/913640.
Ortiz M, Torréns M, Alakulppi N, Strömbom L, Fragoso A, O’Sullivan CK. Amperometric supramolecular genosensor self-assembled on cyclodextrin-modified surfaces. Electrochem Commun. 2011;13:578–81.
Bartolome JP, Echegoyen L, Fragoso A. Reactive carbon nano-onion modified glassy carbon surfaces as DNA sensors for human papillomavirus oncogene detection with enhanced sensitivity. Anal Chem. 2015;87:6744–51.
Zhao Z, Tapec-Dytioco R, Tan W. Unltrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. J Am Chem Soc. 2003;125:11474–5.
Riccò R, Meneghello A, Enrichi F. Signal enhancement in DNA microarray using dye doped silica nanoparticles: application to human papilloma virus (HPV) detection. Biosens Bioelectron. 2011;26:2761–5.
Lee CC, Liao YC, Lai YH, Chuang MC. Recognition of dual targets by a molecular beacon-based sensor: subtyping of influenza A virus. Anal Chem. 2015;87:5410–6.
Li H, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Nat Ac Sci USA. 2004;101:14036–9.
Li J, Song S, Liu X, Wang L, Pan D, Huang Q, Zhao Y. Enzyme-based multi-component optical nanoprobes for sequence-specific detection of DNA hybridization. Adv Mater. 2008;20:497–500.
Ma C, Wang W, Mulchandani A, Shi C. A simple colorimetric DNA detection by target-induced hybridization chain reaction for isothermal signal amplification. Anal Biochem. 2014;457:19–23.
Lu S, Hu T, Wang S, Sun J, Yang X. Ultra-sensitive colorimetric assay system based on the hybridization chain reaction-triggered enzyme cascade amplification. ACS Appl Mater Interfaces. 2017;9:167–75.
Sun J, Ge J, Liu W, Lan M, Zhang H, Wang P, Wang Y, Niu Z. Multi-enzyme co-embedded organic-inorganic hybrid nanoflowers: synthesis and application as a colorimetric sensor. Nano. 2014;6:255–62.
Fu J, Liu M, Liu Y, Woodbury NW, Yan H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J Am Chem Soc. 2012;134:5516–9.
Müller J, Niemeyer CM. DNA-directed assembly of artificial multienzyme complexes. Biochem Biophys Res Commun. 2008;377:62–7.
Acknowledgements
This work was supported financially by the FP7-PEOPLE-2011-CIG DeCoDeB project grant awarded to LM. GBA acknowledges the Universitat Rovira i Virgili for the doctoral fellowship. The authors thank Dr. Gert Folkers (Utrecht University, Netherlands) for the generous gift of the construct containing the LacI headpiece domain gene.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 569 kb)
Rights and permissions
About this article
Cite this article
Aktas, G.B., Skouridou, V. & Masip, L. Nucleic acid sensing with enzyme-DNA binding protein conjugates cascade and simple DNA nanostructures. Anal Bioanal Chem 409, 3623–3632 (2017). https://doi.org/10.1007/s00216-017-0304-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0304-z