Abstract
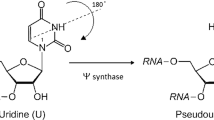
Pseudouridylation affects almost all types of RNAs and the malfunction of pseudouridine synthases, the enzymes responsible for the uridine–pseudouridine transformation, is linked to severe diseases, like cancer and X-linked dyskeratosis congenita. Stand-alone and guide-dependent pseudouridine synthases share a common active site structure and are assumed to share the catalytic mechanism whose details are not yet elucidated. We performed quantum chemical calculations on model systems to investigate the initial steps of several pathways proposed in the literature or based on biochemical analogy and chemical intuition. Results suggest that the Michael addition scheme is unlikely since no stable adduct is formed between the C6-atom of the uridine and the catalytic aspartate. The nucleophilic substitution scheme is ruled out owing to the unfavorable steric arrangement of the reactants. Our results are in favor of the glycal scheme and provide details for the mechanism that is likely to start with the glycosidic bond cleavage between the ribose and uracil, followed by or coupled to the deprotonation of the C2′-atom of the sugar by the conserved catalytic aspartate. A possible role of the latter step is suggested to be the regulation of the intermediate reactivity: C2′ deprotonation leads to a low-energy intermediate with sufficient lifetime to allow base repositioning before reattachment to ribose by C–C bond formation.










Similar content being viewed by others
References
Machnicka MA et al (2013) MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res 41:262–267
Kellner S et al (2014) Profiling of RNA modifications by multiplexed stable isotope labelling. Chem Commun 50:3516–3518
Davis F, Allen FW (1957) Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem 227:907–915
Cohn WE (1959) 5-Ribosyl uracil, a carbon–carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta 32:569–571
Ge J, Yu Y (2013) RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 38:210–218
Liang X-H, Liu Q, Fournier MJ (2009) Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15:1716–1728
Rintala-Dempsey AC, Kothe U (2017) Eukaryotic stand-alone pseudouridine synthases—RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14:1185–1196
Ferré-D’Amaré AR (2003) RNA-modifying enzymes. Curr Opin Struct Biol 13:49–55
Boschi-Muller S, Motorin Y (2013) Chemistry enters nucleic acids biology: enzymatic mechanisms of RNA modification. Biochemistry (Mosc) 78:1392–1404
Yu Y, Meier UT (2014) RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA Biol 11:1483–1494
Baker DL et al (2005) RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev 19:1238–1248
Charpentier B, Muller SB, Branlant C (2005) Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 33:3133–3144
Heiss NS et al (1998) X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19:32–38
Kirwan M, Dokal I (2009) Dyskeratosis congenita, stem cells and telomeres. Biochim Biophys Acta 1792:371–379
Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N (2004) Report missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 74:1303–1308
Penzo M, Guerrieri AN, Zacchini F, Treré D, Montanaro L (2017) RNA pseudouridylation in physiology and medicine: for better and for worse. Genes (Basel) 8:301
Kierzek E et al (2014) The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42:3492–3501
Meroueh M et al (2000) Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res 28:2075–2083
Charette M, Gray MW (2000) Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49:341–351
Hamma T, Ferré-D’Amaré AR (2006) Pseudouridine synthases. Chem Biol 13:1125–1135
Liang B, Li H (2011) Structures of ribonucleoprotein particle modification enzymes. Q Rev Biophys 44:95–122
Majumder M, Bosmeny MS, Gupta R (2016) Structure–function relationships of archaeal Cbf5 during in vivo RNA-guided pseudouridylation. RNA 22:1604–1619
Spenkuch F, Motorin Y, Helm M (2014) Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11:1540–1554
Spenkuch F (2013) Open questions how do pseudouridine synthases work? JUnQ Open Quest 3:16–20
Gu X, Liu Y, Santi DV (1999) The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. PNAS 96:14270–14275
Friedt J, Leavens FMV, Mercier E, Wieden H, Kothe U (2014) An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. Nucleic Acids Res 42:3857–3870
Huang L, Pookanjanatavip M, Gu X, Santi DV (1998) A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 2960:344–351
Meier UT (2005) The many facets of H/ACA ribonucleoproteins. Chromosoma 114:1–14
Miracco EJ, Mueller EG (2011) The products of 5-fluorouridine by the action of the pseudouridine synthase TruB disfavor one mechanism and suggest another. J Am Chem Soc 133:11826–11829
Spenkuch F et al (2014) Dye label interference with RNA modification reveals 5-fluorouridine as non-covalent inhibitor. Nucleic Acids Res 42:12735–12745
Spedaliere CJ, Mueller EG (2004) Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 10:192–199
Zhou J, Liang B, Li H (2010) Functional and structural impact of target uridine substitutions on the H/ACA ribonucleoprotein particle pseudouridine synthase. Biochemistry 49:6276–6281
Zhou J et al (2010) Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study. J Mol Biol 401:690–695
Czudnochowski N et al (2014) The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs. Nucleic Acids Res 42:2037–2048
Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG (2004) The pseudouridine synthases: revisiting a mechanism that seemed settled. J Am Chem Soc 126:12758–12759
McDonald MK, Miracco EJ, Chen J, Xie Y, Mueller EG (2011) The handling of the mechanistic probe 5-fluorouridine by the pseudouridine synthase TruA and its consistency with the handling of the same probe by the pseudouridine synthases TruB and RluA. Biochemistry 50:426–436
Veerareddygari GR, Singh SK, Mueller EG (2016) The pseudouridine synthases proceed through a glycal intermediate. J Am Chem Soc 138:7852–7855
Lee T, Kollman PA (1999) Quantum mechanical calculations of nucleophilic attack in the pseudouridine synthesis reaction. J Am Chem Soc 121:9928–9931
Liang B et al (2009) Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16:740–746
Duan J, Li L, Lu J, Wang W, Ye K (2009) Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34:427–439
Pronk S et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Aliev AE et al (2014) Motional timescale predictions by molecular dynamics simulations: case study using proline and hydroxyproline sidechain dynamics. Proteins 82:195–215
Steinbrecher T, Latzer J, Case DA (2012) Revised AMBER parameters for bioorganic phosphates. J Chem Theory Comput 8:4405–4412
Bergonzo C, Cheatham TE (2015) Improved force field parameters lead to a better description of RNA structure. J Chem Theory Comput 11:3969–3972
Vangaveti S, Ranganathan SV, Chen AA (2017) Advances in RNA molecular dynamics: a simulator’s guide to RNA force fields. Wiley Interdiscip Rev RNA 8:e1396
Tan D, Piana S, Dirks RM, Shaw DE (2018) RNA force field with accuracy comparable to state-of-the-art protein force fields. PNAS 115:E1346–E1355
Izadi S, Anandakrishnan R, Onufriev AV (2014) Building water models: a different approach. J Phys Chem Lett 5:3863–3871
Becke A (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Hehre WJ, Ditchfield R, Pople JA (1972) Self-consistent molecular orbital methods. XII. Further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J Chem Phys 56:2257–2261
Grimme S, Antony J, Ehrlich S, Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys 132:154104
Bochevarov AD et al (2013) Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem 113:2110–2142
Toulouse J, Zhu W, Savin A, Jansen G, Ángyán JG (2011) Closed-shell ring coupled cluster doubles theory with range separation applied on weak intermolecular interactions. J Chem Phys 135:084119
Heßelmann A (2012) Random-phase-approximation correlation method including exchange interactions. Phys Rev A 85:012517
Kállay M (2015) Linear-scaling implementation of the direct random-phase approximation. J Chem Phys 142:204105
MRCC, a quantum chemical program suite written by M. Kállay, Z. Rolik, J. Csontos, P. Nagy, G. Samu, D. Mester, J. Csóka, B. Szabó, I. Ladjánszki, L. Szegedy, B. Ladóczki, K. Petrov, M. Farkas, P. D. Mezei, and B. Hégely. See also Z. Rolik, L. Szegedy, I. Ladjánszki, B. Ladóczki, and M. Kállay, J Chem Phys 139:094105 (2013). https://www.mrcc.hu
Jacobs AL, Schär P (2012) DNA glycosylases: in DNA repair and beyond. Chromosoma 121:1–20
Dinner AR, Blackburn GM, Karplus M (2001) Uracil-DNA glycosylase acts by substrate autocatalysis. Nature 413:752–755
Cavallo L, Kleinjung J, Fraternali F (2003) POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acid Res 31:3364–3366
Acknowledgements
This work was supported by the Hungarian Scientific Research Fund (OTKA) through Grants K111862 and K116305 and by the MedInProt Protein Science Research Synergy Program. J.O. was supported by the Bolyai János Research Scholarship and by NKFIH Grant No. 115503. Part of the computations were performed using the supercomputing service offered by the Hungarian National Information Infrastructure Development Institute. This work is dedicated to the memory of Professor János G. Ángyán.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published as part of the special collection of articles In Memoriam of János Ángyán.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kiss, D.J., Oláh, J., Tóth, G. et al. Quantum chemical calculations support pseudouridine synthase reaction through a glycal intermediate and provide details of the mechanism. Theor Chem Acc 137, 162 (2018). https://doi.org/10.1007/s00214-018-2361-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-018-2361-6