Abstract
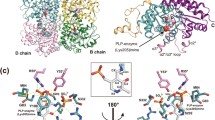
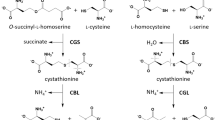
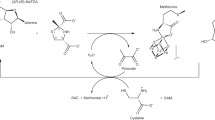
The γ-elimination mechanism of l-methionine catalyzed by methionine γ-lyase was studied by employing the combined quantum mechanics and molecular (QM/MM) calculations. Based on the two crystal structures of methionine γ-lyase from Clostridium sporogenes and Citrobacter freundii, a computational model that contains the enzyme and the external aldimine intermediate was constructed. According to the results of our calculations, the whole catalytic reaction can be divided into two parts. Part I describes the formation of external aldimine intermediate from the internal aldimine intermediate, in which the key protonation step within the tetrahedral intermediate (IM2) should require the assistance of one water molecule, and the dissociation of Lys210 from the internal aldimine intermediate corresponds to an overall energy barrier of 13.5 kcal/mol. In Part II, the external aldimine intermediate converts to the aminocrotonate intermediate, which contains complex asynchronous and concerted proton transfer processes, including those of Cα-proton and Cβ-proton of the substrate as well as the C4′-proton of PLP, in which the pocket residues Tyr113 and Lys210 play key roles. The abstraction of Cβ-proton and the transfer of C4′-proton correspond to very similar barrier heights (16.5 and 16.2 kcal/mol) on the potential energy surface, implying that both steps are possibly rate-limiting. Besides, the calculated overall energy barrier is close to the free energy barrier estimated from the experiment (16.3 kcal/mol). These results may provide useful information for further understanding the catalysis of pyridoxal 5′-phosphate (PLP)-dependent lyase.












Similar content being viewed by others
References
Tanaka H, Esaki N, Soda K (1985) A versatile bacterial enzyme: l-methionine γ-lyase. Enzyme Microb Technol 7:530–537
Tanaka H, Esaki N, Soda K (1977) Properties of l-methionine γ-lyase from Pseudomonas ovalis. Biochemistry 16:100–106
Manukhov IV, Mamaeva DV, Rastorguev SM, Faleev NG, Morozova EA, Demidkina TV, Zavilgelsky GB (2005) A gene encoding l-methionine γ-lyase is present in Enterobacteriaceae family genomes: identification and characterization of Citrobacter freundii l-methionine γ-lyase. J Bacteriol 187:3889–3893
Nakayama T, Esaki N, Lee WJ, Tanaka I, Tanaka H, Soda K (1984) Purification and properties of l-methionine γ-lyase from Aeromonas sp. Agric Biol Chem 48:2367–2369
Kreis W, Hession C (1973) Isolation and purification of l-methionine-α-deamino-γ-mercaptomethane-lyase (L-methioninase) from Clostridium sporogenes. Cancer Res 33:1862–1865
Yoshimura M, Nakano Y, Yamashita Y, Oho T, Saito T, Koga T (2000) Formation of Methyl Mercaptan from l-Methionine by Porphyromonas gingivalis. Infect Immun 68:6912–6916
Fukamachi H, Nakano Y, Okano S, Shibata Y, Abiko Y, Yamashita Y (2005) High production of methyl mercaptan by L-methionine-α-deamino-γ-mercaptomethane lyase from Treponema denticola. Res Commun 331:127–131
Tokoro M, Asai T, Kobayashi S, Takeuchi T, Nozaki T (2003) Identification and characterization of two isoenzymes of methionine γ-lyase from Entamoeba histolytica. J Biol Chem 278:42717–42727
Lockwood BC, Coombs GH (1991) Purification and characterization of methionine γ-lyase from Trichomonas vaginalis. Biochem J 279:675–682
El-Sayed AS (2010) Microbial L-methioninase: production, molecular characterization, and therapeutic applications. Appl Microbiol Biotechnol 86:445–467
Rébeillé F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103:15687–15692
Alexander W, Sandmeier E, Mehta PK, Christen P (1994) Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Eur J Biochem 219:953–960
Hoffman RM (1997) Methioninase: a therapeutic for diseases related to altered methionine metabolism and transmethylation: cancer, heart disease, obesity, aging, and Parkinson’s disease. Hum Cell 10:69–80
Sato D, Nozaki T (2009) Methionine gamma-lyase: the unique reaction mechanism, physiological roles, and therapeutic applications against infectious diseases and cancers. IUBMB Life 61:1019–1028
Tan Y, Xu M, Hoffman RM (2010) Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res 30:1041–1046
Revtovich S, Anufrieva N, Morozova E, Kulikova V, Nikulin A, Demidkina T (2016) Structure of methionine γ-lyase from Clostridium sporogenes. Acta Cryst 72:65–71
Revtovich SV, Faleev NG, Morozova EA, Anufrieva NV, Nikulin AD, Demidkina TV (2014) Crystal structure of the external aldimine of Citrobacter freundii methionine γ-lyase with glycine provides insight in mechanisms of two stages of physiological reaction and isotope exchange of α-and β-protons of competitive inhibitors. Biochimie 101:161–167
Brzovic P, Holbrook EL, Greene RC, Dunn MF (1990) Reaction mechanism of Escherichia coli cystathionine. gamma.-synthase: direct evidence for a pyridoxamine derivative of vinylgloxylate as a key intermediate in pyridoxal phosphate dependent. gamma.-elimination and. gamma.-replacement reactions. Biochemistry 29:442–451
Anufrieva NV, Faleev NG, Morozova EA, Bazhulina NP, Revtovich SV, Timofeev VP, Tkachev YV, Nikulin AD, Demidkina TV (2015) The role of active site tyrosine 58 in Citrobacter freundii methionine γ-lyase. Biochim Biophys Acta 1854:1220–1228
Kuznetsov NA, Faleev NG, Kuznetsova AA, Morozova EA, Revtovich SV, Anufrieva NV, Nikulin AD, Fedorova OS, Demidkina TV (2015) Pre-steady-state kinetic and structural analysis of interaction of methionine γ-lyase from Citrobacter freundii with inhibitors. J Biol Chem 1:671–681
Dennington R II, Keith T, Millam J (2009) GaussView, version 5. Semichem Inc., Shawnee Mission
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) GAUSSIAN 03, revision D.01. Gaussian Inc., Wallingford
Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct Funct Genet 61:704–721
Bas DC, Rogers DM, Jensen JH (2008) Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins Struct Funct Genet 73:765–783
Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35:W522–W525
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217
Brooks BR, Brooks CL III, MacKerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614
Chen JH, Im WP, Brooks CL (2006) Balancing solvation and intramolecular interactions: toward a consistent generalized Born force field. J Am Chem Soc 128:3728–3736
MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616
Ahlrichs R, Bar M, Haser M, Horn H, Kolmel C (1989) Electronic structure calculations on workstation computers: the program system turbomole. Chem Phys Lett 162:165–169
Smith W, Forester TR (1996) A general-purpose parallel molecular dynamics simulation package. J Mol Graphics 14:136–141
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38:3098–3100
Becke AD (1993) A new mixing of Hartree–Fock and local density-functional theories. J Chem Phys 98:1372–1377
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Bakowies D, Thiel W (1996) Hybrid models for combined quantum mechanical and molecular mechanical approaches. J Phys Chem 100:10580–10594
de Vries AH, Sherwood P, Collins SJ, Rigby AM, Rigutto M, Kramer GJ (1999) Zeolite structure and reactivity by combined quantum-chemical–classical calculations. J Phys Chem B 103:6133–6141
Field MJ, Bash PA, Karplus M (1990) A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J Comput Chem 11:700–733
Billeter SR, Turner AJ, Thiel W (2000) Linear scaling geometry optimisation and transition state search in hybrid delocalised internal coordinates. Phys Chem Chem Phys 2:2177–2186
Nocedal J (1980) Updating quasi-Newton matrices with limited storage. Math Comput 35:773–782
Banerjee A, Adams N, Simons J, Shepard R (1985) Search for stationary points on surfaces. J Phys Chem 89:52–57
Sherwood P, de Vries AH, Guest MF, Schreckenbach G, Catlow CRA, French SA, Sokol AA, Bromley ST, Thiel W, Turner AJ, Billeter S, Terstegen F, Thiel S, Kendrick J, Rogers SC, Casci J, Watson M, King F, Karlsen E, Sjøvoll M, Fahmi A, Schafer A, Lennartz C (2003) QUASI: a general purpose implementation of the QM/MM approach and its application to problems in catalysis. J Mol Struct THEOCHEM 632:1–28
Cassimjee KE, Manta B, Himo F (2015) A quantum chemical study of the ω-transaminase reaction mechanism. Org Biomol Chem 13:8453–8464
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21573127, 21373125, 21773138).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
214_2017_2140_MOESM1_ESM.docx
Supplementary material 1. Solvation model for the MD simulation; RMSDs for the protein backbone atoms along the MD trajectory; distances of some important hydrogen bonds in the MD simulation; superimposition of active sites of QM/MM optimized geometries of enzyme–substrate complexes; calculated QM/MM energy profile; optimized geometries of active site of IM2. (DOCX 1911 kb)
Rights and permissions
About this article
Cite this article
Lin, B., Tian, G. & Liu, Y. Mechanistic insights into the γ-elimination reaction of l-methionine catalyzed by methionine γ-lyase (MGL). Theor Chem Acc 136, 105 (2017). https://doi.org/10.1007/s00214-017-2140-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-017-2140-9