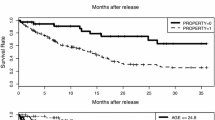
Abstract
Many medical studies aim to identify factors associated with a time to an event such as survival time or time to relapse. Often, in particular, when binary variables are considered in such studies, interactions of these variables might be the actual relevant factors for predicting, e.g., the time to recurrence of a disease. Testing all possible interactions is often not possible, so that procedures such as logic regression are required that avoid such an exhaustive search. In this article, we present an ensemble method based on logic regression that can cope with the instability of the regression models generated by logic regression. This procedure called survivalFS also provides measures for quantifying the importance of the interactions forming the logic regression models on the time to an event and for the assessment of the individual variables that take the multivariate data structure into account. In this context, we introduce a new performance measure, which is an adaptation of Harrel’s concordance index. The performance of survivalFS and the proposed importance measures is evaluated in a simulation study as well as in an application to genotype data from a urinary bladder cancer study. Furthermore, we compare the performance of survivalFS and its importance measures for the individual variables with the variable importance measure used in random survival forests, a popular procedure for the analysis of survival data. These applications show that survivalFS is able to identify interactions associated with time to an event and to outperform random survival forests.



Similar content being viewed by others
References
Alpaydin E (2014) Introduction to machine learning. MIT Press, Cambridge
An P, Feitosa M, Ketkar S, Adelman A, Lin S, Borecki I, Province M (2009) Epistatic interactions of CDKN2B-TCF7L2 for risk of type 2 diabetes and of CDKN2B-JAZF1 for triglyceride/high-density lipoprotein ratio longitudinal change: evidence from the Framingham Heart Study. BMC Proc 3:S71
Andrew AS, Karagas MR, Nelson HH, Guarrera S, Polidoro S, Gamberini S, Sacerdote C, Moore JH, Kelsey KT, Demidenko E, Vineis P, Matullo G (2008) DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. Hum Hered 65:105–118
Banerjee M, Filson C, Xia R, Miller DC (2014) Logic regression for provider effects on kidney cancer treatment delivery. Comput Math Methods Med 2014:316,935
Bender R, Augustin T, Blettner M (2005) Generating survival times to simulate Cox proportional hazards models. Tech. Rep. 11, Stat Med
Bivard A, Levi C, Lin L, Cheng X, Aviv R, Spratt NJ, Lou M, Kleinig T, O’Brien B, Butcher K, Zhang J, Jannes J, Dong Q, Parsons M (2017) Validating a predictive model of acute advanced imaging biomarkers in ischemic stroke. Stroke 48(3):645–650
Bowers K, Li Q, Bressler J, Avramopoulos D, Newschaffer C, Fallin MD (2011) Glutathione pathway gene variation and risk of autism spectrum disorders. J Neurodev Disord 3(2):132–143
Breiman L (1996) Bagging predictors. Mach Learn 24(2):123–140
Breslow N (1974) Covariance analysis of censored survival data. Biometrics 30(1):89–99
Buehlmann P, Yu B (2002) Analyzing bagging. Ann Stat 30(4):927–961
Carty CL, Heagerty P, Heckbert SR, Jarvik GP, Lange LA, Cushman M, Tracy RP, Reiner AP (2010) Interaction between fibrinogen and IL-6 genetic variants and associations with cardiovascular disease risk in the cardiovascular health study. Ann Hum Genet 74:1–10
Chi C, Street WN, Wohlberg WH (2007) Application of artificial neural network-based survival analysis on two breast cancer datasets. AMIA Ann Symp Proc 30:130–134
Cox DR (1972) Regression models and life tables. J R Stat Soc B 34(2):187–220
Cox DR (1975) Partial likelihood. Biometrika 62(2):269–279
Dazard JE, Ishwaran H, Mehlotra R, Weinberg A, Zimmerman P (2018) Ensemble survival tree models to reveal pairwise interactions of variables with time-to-events outcomes in low-dimensional setting. Stat Appl Genet Mol Biol. https://doi.org/10.1515/sagmb-2017-0038
Dinu I, Mahasirimongkol S, Liu Q, Yanai H, Sharaf Eldin N, Kreiter E, Wu X, Jabbari S, Tokunaga K, Yasui Y (2012) Snp-snp interactions discovered by logic regression explain Crohn’s disease genetics. PLoS One 7(10):e43,035
duVerle DA, Takeuchi I, Murakami-Tonami Y, Kodamatsu K, Tsuda K (2013) Discovering combinatorial interactions in survival data. Bioinformatics 29:3053–3059
Eliot M, Azzoni L, Firnhaber C, Stevens W, Glencross DK, Sanne I, Montaner LJ, Foulkes AS (2009) Tree-based methods for discovery of association between flow cytometry data and clinical endpoints. Adv Bioinform 2009:235,320
Enquobahrie DA, Smith NL, Bis JC, Carty CL, Rice KM, Lumley T, Hindorff LA, Lemaitre RN, Williams MA, Siscovick DS, Heckbert SR, Psaty BM (2008) Cholesterol ester transfer protein, interleukin-8, peroxisome proliferator activator receptor alpha, and toll-like receptor 4 genetic variations and risk of incident nonfatal myocardial infarction and ischemic stroke. Am J Cardiol 101:1683–1688
Etzioni R, Falcon S, Gann PH, Kooperberg CL, Penson DF, Stampfer MJ (2004) Prostate-specific antigen and free prostate-specific antigen in the early detection of prostate cancer: do combination tests improve detection? Cancer Epidemiol Biomark Prev 13:1640–1645
Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB (2005) Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst 97:273–282
Garte S (2001) Metabolic susceptibility genes as cancer risk factors: time for a reassessment? Cancer Epidemiol Biomark Prev 10:1233–1237
Graf E, Schmoor C, Sauerbrei W, Schumacher M (1999) Assessment and comparison of prognostic classification schemes for survival data. Stat Med 18:2529–2545
Grotenhuis AJ, Dudek AM, VG W, Witjes JA, Aben KK, van der Mare SL, Vermeulen SH, Kiemeney LA (2014) Prognostic relevance of urinary bladder cancer susceptibility loci. PLoS One 9:e89,164
Gui J, Moore JH, Kelsey KT, Marsit CJ, Karagas MR, Andrew AS (2011) A novel survival multifactor dimensionality reduction method for detecting gene-gene interactions with application to bladder cancer prognosis. Hum Genet 129(1):101–110
Harrell F, Califf R, Pryor D, Lee K, Rosati R (1982) Evaluating the yield of medical tests. J Am Med Assoc 247(18):2543–2546
Harth V, Schaefer M, Abel J, Maintz L, Neuhaus T, Besuden M, Primke R, Wilkesmann A, Thier R, Vetter H, Ko YD, Bruening T, Bolt HM, Ickstadt K (2008) Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A 71:887–897
Hirahara N, Fujii Y, Yamamoto T, Hyakudomi R, Hirayama T, Taniura T, Ishitobi K, Tajima Y (2017) Validation of a novel prognostic scoring system using inflammatory response biomarkers in patients undergoing curative thoracoscopic esophagectomy for esophageal squamous cell carcinoma. OncoTargets Ther 10:363–370
Hothorn T, Lausen B, Benner A, Radespiel-Troeger M (2004) Bagging survival trees. Stat Med 23(1):77–91
Huang J, Lin A, Narasimhan B, Quertermous T, Hsiung CA, Ho LT, Grove JS, Olivier M, Ranade K, Risch NJ, Olshen RA (2004) Tree-structured supervised learning and the genetics of hypertension. Proc Natl Acad Sci USA 101:10,529–10,534
Ickstadt K, Schaefer M, Fritsch A, Schwender H, Abel J, Bolt HM, Bruening T, Ko YD, Vetter H, Harth V (2008) Statistical methods for detecting genetic interactions: a head and neck squamous-cell cancer study. J Toxicol Environ Health A 71:803–815
Ishwaran H (2007) Variable importance in binary regression trees and forests. Electron J Stat 1:519–537
Ishwaran H, Kogalur UB (2007) Random survival forests for R. Rnews 7(2):25–31
Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS (2008) Random survival forests. Ann Appl Stat 2(3):841–860
Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS (2010) High-dimensional variable selection for survival data. J Am Stat Assoc 105:205–217
Janes H, Pepe M, Kooperberg C, Newcomb P (2005) Identifying target populations for screening or not screening using logic regression. Stat Med 24:1321–1338
Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, Rabstein S, Selinski S, Mueller T, Ickstadt K, Gilbert M, Ko YD, Baisch C, Pesch B, Harth V, Bolt HM, Vollmert C, Illig T, Eils R, Dippon J, Brauch H (2008) Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat 108:137–149
Keles S, van der Laan MJ, Vulpe C (2004) Regulatory motif finding by logic regression. Bioinformatics 20:2799–2811
Klein JP, Moeschberger ML (1997) Survival analysis. Springer, New York
Kooperberg C, Bis JC, Marciante KD, Heckbert SR, Lumley T, Psaty BM (2007) Logic regression for analysis of the association between genetic variation in the renin–angiotensin system and myocardial infarction or stroke. Am J Epidemiol 165:334–343
Lee S, Kwon MS, Oh JM, Park T (2012) Gene-gene interaction analysis for the survival phenotype based on the Cox model. Bioinformatics 28(18):i582–i588
Li Q, Fallin MD, Louis TA, Lasseter VK, McGrath JA, Avramopoulos D, Wolyniec PS, Valle D, Liang KY, Pulver AE, Ruczinski I (2010) Detection of SNP–SNP interactions in trios of parents with schizophrenic children. Genet Epidemiol 34(5):396–406
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K (2000) Environmental and heritable factors in the causation of cancer, analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343(2):78–85
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD (2007) A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet 80(6):1125–1137
Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, St Clair D, Muglia P, Shugart YY, Weinberger DR (2010) Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet 127:441–452
Park M, Hastie T (2007) \(L_1\)-regularization path algorithm for generalized linear models. J R Stat Soc B 69:659–677
Poole EM, Hsu L, Xiao L, Kulmacz RJ, Carlson CS, Rabinovitch PS, Makar KW, Potter JD, Ulrich CM (2010) Genetic variation in prostaglandin E2 synthesis and signaling, prostaglandin dehydrogenase, and the risk of colorectal adenoma. Cancer Epidemiol Biomark Prev 19:547–557
Raimondi S, Gandini S, Fargnoli MC, Bagnardi V, Maisonneuve P, Specchia C, Kumar R, Nagore E, Han J, Hansson J (2012) Melanocortin-1 receptor, skin cancer and phenotypic characteristics (M-SKIP) project: study design and methods for pooling results of genetic epidemiological studies. BMC Med Res Methodol 12(1):116
Rathod SD, Li T, Klausner JD, Hubbard A, Reingold AL, Madhivanan P (2015) Logic regression-derived algorithms for syndromic management of vaginal infections. BMC Med Inform Decis Mak 15(1):106
Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH (2001) Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 69(1):138–147
Ruczinski I, Kooperberg C, LeBlanc M (2003) Logic regression. J Comput Graph Stat 12:475–511
Ruczinski I, Kooperberg C, LeBlanc M (2004) Exploring interactions in high-dimensional genomic data: an overview of logic regression, with applications. J Mult Anal 90:178–195
Sapkota Y, Mackey JR, Lai R, Franco-Villalobos C, Lupichuk S, Robson PJ, Kopciuk K, Cass CE, Yasui Y, Damaraju S (2014) Assessing SNP–SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS One 8(6):e64,896
Sarbakhsh P, Mehrabi Y, Daneshpour MS, Zayeri F, Zarkesh M (2013) Logic regression analysis of association of gene polymorphisms with low HDL: Tehran lipid and glucose study. Gene 513(2):278–281
Schwender H, Ickstadt K (2008) Identification of SNP interactions using logic regression. Biostatistics 9:187–198
Schwender H, Ruczinski I (2010) Logic regression and its extensions. In: Dunlap JC, Moore JH (eds) Computational methods for genetics of complex traits. Band 72 von advances in genetics. Academic Press, Amsterdam, pp 25–45
Schwender H, Bowers K, Fallin MD, Ruczinski I (2011a) Importance measures for epistatic interactions in case-parent trios. Ann Hum Genet 75:122–132
Schwender H, Ruczinski I, Ickstadt K (2011b) Testing SNPs and sets of SNPs for importance in association studies. Biostatistics 12:18–32
Segal MR, Barbour JD, Grant RM (2004) Relating HIV-1 sequence variation to replication capacity via trees and forests. Stat Appl Genet Mol Biol 3:2
Seki S, Fujiwara M, Matsuura M, Fujita S, Ikeda H, Asahina I, Ikeda T (2011) Prediction of outcome of patients with oral squamous cell carcinoma using vascular invasion and the strongly positive expression of vascular endothelial growth factors. Oral Oncol 47(7):588–593
Selinski S (2014) Urinary bladder cancer risk variants: recent findings and new challenges of GWAS and confirmatory studies. Arch Toxicol 88(7):1469–1475
Selinski S, Bürger H, Blaszkewicz M, Otto T, Volkert F, Moormann O, Niedner H, Hengstler GJ, Golka K (2016) Occupational risk factors for relapse-free survival in bladder cancer patients. J Toxicol Environ Health A 79:1136–1143
Sharafeldin N, Slattery ML, Liu Q, Franco-Villalobos C, Caan BJ, Potter JD, Yasui Y (2015) A candidate-pathway approach to identify gene–environment interactions: analyses of colon cancer risk and survival. J Natl Cancer Inst. https://doi.org/10.1093/jnci/djv160
Su X, Zhou T, Yan X, Fan J, Yang S (2008) Interaction trees with censored survival data. Int J Biostat 4(1):2
Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan ASY, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JPJ, Yuen ST, Leung SY, Hamilton SR (2008) Epigenetic–genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res 14:2560–2569
Vaidya VS, Waikar SS, Ferguson MA, Collings FB, Sunderland K, Gioules C, Bradwin G, Matsouaka R, Betensky R, Curhan GC, Bonventre JV (2008) Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin Transl Sci 3:200–208
Van Belle V, Pelckmans K, van Huffel S, Suykens JA (2011) Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif Intell Med 53:107–118
Van Rhijn BW, Catto JW, Goebell PJ, Knuechel R, Shariat SF, van der Poel HG, Sanchez-Carbayo M, Thalmann GN, Schmitz-Draeger BJ, Kiemeney LA (2014) Molecular markers for urothelial bladder cancer prognosis: toward implementation in clinical practice. Urol Oncol 32:1078–1087
Wang MH, Fiocchi C, Ripke S, Zhu X, Duerr RH, Achkar JP (2013) A novel approach to detect cumulative genetic effects and genetic interactions in Crohn’s disease. Inflamm Bowel Dis 19(9):1799–1808
Wang MH, Fiocchi C, Zhu X, Ripke S, Kamboh MI, Rebert N, Duerr RH, Achkar JP (2014) Gene–gene and gene–environment interactions in ulcerative colitis. Hum Genet 133(5):547–558
Wright MN, Ziegler A, König IR (2016) Do little interactions get lost in dark random forests? BMC Bioinform 17(1):145
Xu H, Liu R, He B, Bi CW, Bi K, Li Q (2016) Polyamine metabolites profiling for characterization of lung and liver cancer using an LC-tandem MS method with multiple statistical data mining strategies: discovering potential cancer biomarkers in human plasma and urine. Molecules 21(8):1040
Yaziji H, Battifora H, Barry TS, Hwang HC, Bacchi CE, McIntosh MW, Kussick SJ, Gown AM (2006) Evaluation of 12 antibodies for distinguishing epithelioid mesothelioma from adenocarcinoma: identification of a three-antibody immunohistochemical panel with maximal sensitivity and specificity. Mod Pathol 19:514–523
Zhi S, Li Q, Yasui Y, Edge T, Topp E, Neumann NF (2015) Assessing host-specificity of escherichia coli using a supervised learning logic-regression-based analysis of single nucleotide polymorphisms in intergenic regions. Mol Phylogenet Evol 92:72–81
Acknowledgements
The authors would like to thank Hannah Bürger for help with the simulation study. This work was supported by the Deutsche Forschungsgemeinschaft (SCHW 1508/3-1 to H.S.; project C4 of the Collaborative Research Center SFB 876 to K.I.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tietz, T., Selinski, S., Golka, K. et al. Identification of interactions of binary variables associated with survival time using survivalFS. Arch Toxicol 93, 585–602 (2019). https://doi.org/10.1007/s00204-019-02398-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-019-02398-6