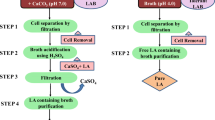
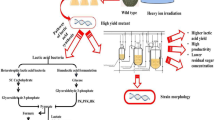
Abstract
Lactic acid bacteria (LAB) can take advantage of fermentable carbohydrates to produce lactic acid. They are proverbially applied in industry, agricultural production, animal husbandry, food enterprise, pharmaceutical engineering and some other important fields, which are closely related to human life. For performing the probiotic functions, LAB have to face the low pH environment of the gastrointestinal tract. Therefore, acid resistance of LAB is of great importance not only for their own growth, but also for fermentation and preparation of probiotic products. Recent research studies on acid resistance mechanisms of LAB are mainly focused on neutralization process, biofilm and cell density, proton pump, protection of macromolecules, pre-adaptation and cross-protection, and effect of solutes. In this context, biotechnological strategies such as synthetic biology, genome shuffling, high pressure homogenization and adaptive laboratory evolution were also used to improve the acid resistance of LAB to respond to constantly changing low pH environment.

Similar content being viewed by others
References
Abdullah AlM, Sugimoto S, Higashi C, Matsumoto S, Sonomoto K (2010) Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microbiol 76(13):4277–4285
Arena ME, Saguir FM, de Nadra MC (1999) Arginine dihydrolase pathway in Lactobacillus plantarum from orange. Int J Food Microbiol 47:203–209
Arena ME, de Nadra MC, Muñoz R (2002) The arginine deiminase pathway in the wine lactic acid bacterium Lactobacillus hilgardii X 1 B: structural and functional study of the arcABC genes. Gene 301:61–66
Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR (2004) Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microb 70(9):5315–5322
Bravo-Ferrada B, Hollmann A, Delfederico L, Valdés La Hens D, Caballero A, Semorile L (2013) Patagonian red wines: selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J Microbiol Biotechnol 29(9):1537–1549
Broadbent JR, Larsen RL, Deibel V, Steele JL (2010) Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J Bacteriol 192(9):2445–2458
Cappa F, Cattivelli D, Cocconcelli PS (2005) The uvrA gene is involved in oxidative and acid stress responses in Lactobacillus helveticus CNBL1156. Res Microbiol 156:1039–1047
Cardoso FS, Gaspar P, Hugenholtz J, Ramos A, Santos H (2004) Enhancement of trehalose production in dairy propionibacteria through manipulation of environmental conditions. Int J Food Microbiol 91(2):195–204
Carvalho AL, Cardoso FS, Bohn A, Neves AR, Santos H (2011) Engineering trehalose synthesis in Lactococcus lactis for improved stress tolerance. Appl Environ Microbiol 77(12):4189–4199
Champomier MC, Zúñiga M, Morel-Deville F, Pérez-Martínez G, Zagorec M, Ehrlich SD (1999) Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol Lett 180:297–304
Corcoran BM, Stanton C, Fitzgerald GF, Ross RP (2007) Growth of probiotic lactobacilli in the presence of oleic acid enhances subsequent survival in gastric juice. Microbiology 153(1):291–299
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745
Cotter PD, Hill C (2003) Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol R 67(3):429–453
Cueva OA (2009) Pulsed electric field influences on acid tolerance, bile tolerance, protease activity and growth characteristics of Lactobacillus acidophilus La-k. Dissertation, Louisiana State University
De Angelis M, Gobbetti M (2004) Environmental stress responses in Lactobacillus: a review. Proteomics 4(1):106–122
De Angelis M, Di Cagno R, Huet C, Crecchio C, Fox PF, Gobbetti M (2004) Heat shock response in Lactobacillus plantarum. Appl Environ Microbiol 70(3):1336–1346
Feehily C, Karatzas KAG (2013) Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol 114(1):11–24
Fernandez A, Ogawa J, Penaud S, Boudebbouze S, Ehrlich D, van de Guchte M, Maguin E (2008) Rerouting of pyruvate metabolism during acid adaptation in Lactobacillus bulgaricus. Proteomics 8(15):3154–3163
Gaspar P, Carvalho AL, Vinga S, Santos H, Neves AR (2013) From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria. Biotechnol Adv 31(6):764–788
Hall-Stoodley L, Stoodley P (2009) Evolving concepts in biofilm infections. Cell Microbiol 11(7):1034–1043
Hartke A, Bouche S, Laplace J-M, Benachour A, Boutibonnes P, Auffray Y (1995) UV-inducible proteins and UV-induced crossprotection against acid, ethanol, H2O2 or heat treatments in Lactococcus lactis subsp. lactis. Arch Microbiol 163:329–336
Heunis T, Deane S, Smit S, Dicks LM (2014) Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J Proteome Res 13(9):4028–4039
Kajfasz JK, Jr RGQ (2011) Responses of lactic acid bacteria to acid stress. In: Tsakalidou E, Papadimitrious K (eds) Stress responses of lactic acid bacteria. Springer, New York, pp 23–53
Kim SG, Batt CA (1993) Cloning and sequencing of the Lactococcus lactis subsp. lactis groESL operon. Gene 127:121–126
Kim JE, Eom HJ, Kim Y, Ahn JE, Kim JH, Han NS (2012) Enhancing acid tolerance of Leuconostoc mesenteroides with glutathione. Biotechnol Lett 34(4):683–687
Koponen J, Laakso K, Koskenniemi K, Kankainen M, Savijoki K, Nyman TA et al (2012) Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J Proteomics 75(4):1357–1374
Kullen MJ, Klaenhammer TR (1999) Identification of the pH-inducible, proton-translocating F1-F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol Microbiol 33(6):1152–1161
Lee K, Lee H, Pi K, Choi Y (2008) The effect of low pH on protein expression by the probiotic bacterium Lactobacillus reuteri. Proteomics 8(8):1624–1630
Liu Y, Tang H, Lin Z, Xu P (2015) Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv 33(7):1484–1492
Lucas PM, Blancato VS, Claisse O, Magni C, Lolkema JS, Lonvaud-Funel A (2007) Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylation operon in a putative acid resistance locus. Microbiology 153(7):2221–2230
Maghnouj A, De Sousa TF, Stalon V, Vander Wauven C (1998) The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J Bacteriol 180:6468–6475
Michelson T, Kask K, Jõgi E, Talpsep E, Suitso I, Nurk A (2006) L(+)-Lactic acid producer Bacillus coagulans SIM-7 DSM 14043 and its comparison with Lactobacillus delbrueckii ssp. lactis DSM 20073. Enzym Microb Technol 39(4):861–867
Muramalla T, Aryana KJ (2011) Some low homogenization pressures improve certain probiotic characteristics of yogurt culture bacteria and Lactobacillus acidophilus LA-K. J Dairy Sci 94(8):3725–3738
Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WPC, Ryan CM, Cardayre S (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Patrignani F, Burns P, Serrazanetti D, Vinderola G, Reinheimer JA, Lanciotti R et al (2009) Suitability of high pressure-homogenized milk for the productionof probiotic fermented milk containing Lactobacillus paracasei and Lactobacillus acidophilus. J Dairy Res 76:74–82
Portnoy VA, Bezdan D, Zengler K (2011) Adaptive laboratory evolution-harnessing the power of biology for metabolic engineering. Curr Opin Biotechnol 22(4):590–594
Solieri L, Genova F, De Paola M, Giudici P (2010) Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. J Appl Microbiol 108(1):285–298
Stefanovic E, Fitzgerald G, McAuliffe O (2017) Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol 61:33–49
Stephanopoulos G (2002) Metabolic engineering by genome shuffling. Nat Biotechnol 20(7):666–668
Sumby KM, Grbin PR, Jiranek V (2014) Implications of new research and technologies for malolactic fermentation in wine. Appl Microbiol Biotechnol 98(19):8111–8132
Tabanelli G, Patrignani F, Vinderola G, Reinheimer JA, Gardini F, Lanciotti R (2013) Effect of sub-lethal high pressure homogenization treatments on the in vitro functional and biological properties of lactic acid bacteria. LWT Food Sci Technol 53(2):580–586
Tian H, Tan J, Zhang L, Gu X, Xu W, Guo X, Luo Y (2012) Increase of stress resistance in Lactococcus lactis via a novel food-grade vector expressing a shsp gene from Streptococcus thermophilus. Braz J Microbiol 43(3):1157–1164
To TMH, Grandvalet C, Tourdot-Maréchal R (2011) Cyclopropanation of membrane unsaturated fatty acids is not essential to the acid stress response of Lactococcuslactis subsp. cremoris. Appl Environ Microbiol 77(10):3327–3334
Tourdot-Marechal R, Fortier LC, Guzzo J, Lee B, Divies C (1999) Acid sensitivity of neomycin-resistant mutants of Oenococcus oeni: a relationship between reduction of ATPase activity and lack of malolactic activity. FEMS Microbiol Lett 178:319–326
Trip H, Mulder NL, Lolkema JS (2012) Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524. J Biol Chem 287(14):11195–11204
Triratna L, Saksono B, Sukmarini L, Suparman A (2011) Genome-shuffling-improved acid tolerance and lactic acid production in Lactobacillus plantarum for commercialization. Microbiol Indones 5(1):4
Walker DC, Girgis HS, Klaenhammer TR (1999) The groESL chaperone operon of Lactobacillus johnsonii. Appl Environ Microbiol 65:3033–3041
Wang YH, Li Y, Pei XL, Yu L, Feng Y (2007) Genome-shuffling improved acid tolerance and l-lactic acidvolumetric productivity in Lactobacillus rhamnosus. J Biotechnol 129:510–515
Wilson CM, Loach D, Lawley B, Bell T, Sims LM, O Toole PW, Zomer A, Tannock GW (2014) Lactobacillus reuteri 100 – 23 modulates urea hydrolysis in the murine stomach. Appl Environ Microbiol 80(19):6104–6113
Wu R, Wang W, Yu D, Zhang W, Li Y, Sun Z et al (2009) Proteomics analysis of Lactobacillus casei Zhang, a new probiotic bacterium isolated from traditional home-made koumiss in Inner Mongolia of China. Mol Cell Proteomics 8(10):2321–2338
Wu R, Zhang W, Sun T, Wu J, Yue X, Meng H et al (2011) Proteomic analysis of responses of a new probiotic bacterium Lactobacillus casei Zhang to low acid stress. Int J Food Microbiol 147(3):181–187
Wu C, Zhang J, Chen W, Wang M, Du G, Chen J (2012a) A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl Microbiol Biot 93(2):707–722
Wu C, Zhang J, Wang M, Du G, Chen J (2012b) Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol 39(7):1031–1039
Wu C, Zhang J, Du G, Chen J (2013a) Aspartate protects Lactobacillus casei against acid stress. Appl Microbiol Biot 97(9):4083–4093
Wu C, Zhang J, Du G, Chen J (2013b) Heterologous expression of Lactobacillus casei RecO improved the multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 during salt stress. Bioresour Technol 143:238–241
Wu C, Huang J, Zhou R (2014) Progress in engineering acid stress resistance of lactic acid bacteria. Appl Microbiol Biot 98(3):1055–1063
Yamamoto N, Masujima Y, Takano T (1996) Reduction of membrane bound ATPase activity in a Lactobacillus helveticus strain with slower growth at low pH. FEMS Microbiol Rev 138:179–184
Yokota A, Amachi S, Ishii S, Tomita F (1995) Acid sensitivity of a mutant of Lactococcus lactis subsp. lactis C2 with reduced membrane bound ATPase activity. Biosci Biotechnol Biochem 59:2004–2007
Zhai Z, Douillard FP, An H, Wang G, Guo X, Luo Y, Hao Y (2014) Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 and functional identification of a novel acid stress-related transcriptional regulator Ldb0677. Environ Microbiol 16(6):1524–1537
Zhang J, Fu RY, Hugenholtz J, Li Y, Chen J (2007) Glutathione protects Lactococcus lactis against acid stress. Appl Environ Microbiol 73(16):5268–5275
Zhang J, Wu C, Du G, Chen J (2012) Enhanced acid tolerance in Lactobacillus casei by adaptive evolution and compared stress response during acid stress. Biotechnol Bioprocess Eng 17(2):283–289
Zhu Y, Zhang YP, Li Y (2009) Understanding the industrial application potential of lactic acid bacteria through genomics. Appl Microbiol Biotechnol 83:597–610
Zhu L, Zhu Y, Zhang Y, Li Y (2012) Engineering the robustness of industrial microbes through synthetic biology. Trends Microbiol 20(2):94–101
Acknowledgements
This work was supported by National Nature Science Foundation of China (Grant no. 31371827; 31471712).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Communicated by Yusuf Akhter.
Rights and permissions
About this article
Cite this article
Wang, C., Cui, Y. & Qu, X. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch Microbiol 200, 195–201 (2018). https://doi.org/10.1007/s00203-017-1446-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1446-2